Variance function
| Part of a series on |
| Regression analysis |
|---|
 |
| Models |
| Estimation |
| Background |
|
|
In statistics, the variance function is a smooth function that depicts the variance of a random quantity as a function of its mean. The variance function is a measure of heteroscedasticity and plays a large role in many settings of statistical modelling. It is a main ingredient in the generalized linear model framework and a tool used in non-parametric regression,[1] semiparametric regression[1] and functional data analysis.[2] In parametric modeling, variance functions take on a parametric form and explicitly describe the relationship between the variance and the mean of a random quantity. In a non-parametric setting, the variance function is assumed to be a smooth function.
Intuition
In a regression model setting, the goal is to establish whether or not a relationship exists between a response variable and a set of predictor variables. Further, if a relationship does exist, the goal is then to be able to describe this relationship as best as possible. A main assumption in linear regression is constant variance or (homoscedasticity), meaning that different response variables have the same variance in their errors, at every predictor level. This assumption works well when the response variable and the predictor variable are jointly normal. As we will see later, the variance function in the Normal setting is constant; however, we must find a way to quantify heteroscedasticity (non-constant variance) in the absence of joint Normality.
When it is likely that the response follows a distribution that is a member of the exponential family, a generalized linear model may be more appropriate to use, and moreover, when we wish not to force a parametric model onto our data, a non-parametric regression approach can be useful. The importance of being able to model the variance as a function of the mean lies in improved inference (in a parametric setting), and estimation of the regression function in general, for any setting.
Variance functions play a very important role in parameter estimation and inference. In general, maximum likelihood estimation requires that a likelihood function be defined. This requirement then implies that one must first specify the distribution of the response variables observed. However, to define a quasi-likelihood, one need only specify a relationship between the mean and the variance of the observations to then be able to use the quasi-likelihood function for estimation.[3] Quasi-likelihood estimation is particularly useful when there is overdispersion. Overdispersion occurs when there is more variability in the data than there should otherwise be expected according to the assumed distribution of the data.
In summary, to ensure efficient inference of the regression parameters and the regression function, the heteroscedasticity must be accounted for. Variance functions quantify the relationship between the variance and the mean of the observed data and hence play a significant role in regression estimation and inference.
Types
The variance function and its applications come up in many areas of statistical analysis. A very important use of this function is in the framework of generalized linear models and non-parametric regression.
Generalized linear model
When a member of the exponential family has been specified, the variance function can easily be derived.[4]:29 The general form of the variance function is presented under the exponential family context, as well as specific forms for Normal, Bernoulli, Poisson, and Gamma. In addition, we describe the applications and use of variance functions in maximum likelihood estimation and quasi-likelihood estimation.
Derivation
The generalized linear model (GLM), is a generalization of ordinary regression analysis that extends to any member of the exponential family. It is particularly useful when the response variable is categorical, binary or subject to a constraint (e.g. only positive responses make sense). A quick summary of the components of a GLM are summarized on this page, but for more details and information see the page on generalized linear models.
A GLM consists of three main ingredients:
- 1. Random Component: a distribution of y from the exponential family, [math]\displaystyle{ E[y \mid X] = \mu }[/math]
- 2. Linear predictor: [math]\displaystyle{ \eta = XB = \sum_{j=1}^p X_{ij}^T B_j }[/math]
- 3. Link function: [math]\displaystyle{ \eta = g(\mu), \mu = g^{-1}(\eta) }[/math]
First it is important to derive a couple key properties of the exponential family.
Any random variable [math]\displaystyle{ \textit{y} }[/math] in the exponential family has a probability density function of the form,
- [math]\displaystyle{ f(y,\theta,\phi) = \exp\left(\frac{y\theta - b(\theta)}{\phi} - c(y,\phi)\right) }[/math]
with loglikelihood,
- [math]\displaystyle{ \ell(\theta,y,\phi)=\log(f(y,\theta,\phi)) = \frac{y\theta - b(\theta)}{\phi} - c(y,\phi) }[/math]
Here, [math]\displaystyle{ \theta }[/math] is the canonical parameter and the parameter of interest, and [math]\displaystyle{ \phi }[/math] is a nuisance parameter which plays a role in the variance. We use the Bartlett's Identities to derive a general expression for the variance function. The first and second Bartlett results ensures that under suitable conditions (see Leibniz integral rule), for a density function dependent on [math]\displaystyle{ \theta, f_\theta() }[/math],
- [math]\displaystyle{ \operatorname{E}_\theta\left[\frac{\partial}{\partial \theta} \log(f_\theta(y)) \right] = 0 }[/math]
- [math]\displaystyle{ \operatorname{Var}_\theta\left[\frac{\partial}{\partial \theta}\log(f_\theta(y))\right]+\operatorname{E}_\theta\left[\frac{\partial^2}{\partial \theta^2} \log(f_\theta(y))\right] = 0 }[/math]
These identities lead to simple calculations of the expected value and variance of any random variable [math]\displaystyle{ \textit{y} }[/math] in the exponential family [math]\displaystyle{ E_\theta[y], Var_\theta[y] }[/math].
Expected value of Y: Taking the first derivative with respect to [math]\displaystyle{ \theta }[/math] of the log of the density in the exponential family form described above, we have
- [math]\displaystyle{ \frac{\partial}{\partial \theta}\log(f(y,\theta,\phi))= \frac{\partial}{\partial \theta}\left[\frac{y\theta - b(\theta)}{\phi} - c(y,\phi)\right] = \frac{y-b'(\theta)}{\phi} }[/math]
Then taking the expected value and setting it equal to zero leads to,
- [math]\displaystyle{ \operatorname{E}_\theta\left[\frac{y-b'(\theta)}{\phi}\right] = \frac{\operatorname{E}_\theta[y]-b'(\theta)}{\phi}=0 }[/math]
- [math]\displaystyle{ \operatorname{E}_\theta[y] = b'(\theta) }[/math]
Variance of Y: To compute the variance we use the second Bartlett identity,
- [math]\displaystyle{ \operatorname{Var}_\theta\left[\frac{\partial}{\partial \theta}\left(\frac{y\theta - b(\theta)}{\phi} - c(y,\phi)\right)\right]+\operatorname{E}_\theta\left[\frac{\partial^2}{\partial \theta^2}\left(\frac{y\theta - b(\theta)}{\phi} - c(y,\phi)\right)\right] = 0 }[/math]
- [math]\displaystyle{ \operatorname{Var}_\theta\left[\frac{y - b'(\theta)} \phi \right] + \operatorname{E}_\theta \left[\frac{-b''(\theta)} \phi \right] = 0 }[/math]
- [math]\displaystyle{ \operatorname{Var}_\theta\left[y\right]=b''(\theta)\phi }[/math]
We have now a relationship between [math]\displaystyle{ \mu }[/math] and [math]\displaystyle{ \theta }[/math], namely
- [math]\displaystyle{ \mu = b'(\theta) }[/math] and [math]\displaystyle{ \theta = b'^{-1}(\mu) }[/math], which allows for a relationship between [math]\displaystyle{ \mu }[/math] and the variance,
- [math]\displaystyle{ V(\theta) = b''(\theta) = \text{the part of the variance that depends on }\theta }[/math]
- [math]\displaystyle{ \operatorname{V}(\mu) = b''(b'^{-1}(\mu)). \, }[/math]
Note that because [math]\displaystyle{ \operatorname{Var}_\theta\left[y\right]\gt 0, b''(\theta)\gt 0 }[/math], then [math]\displaystyle{ b': \theta \rightarrow \mu }[/math] is invertible. We derive the variance function for a few common distributions.
Example – normal
The normal distribution is a special case where the variance function is a constant. Let [math]\displaystyle{ y \sim N(\mu,\sigma^2) }[/math] then we put the density function of y in the form of the exponential family described above:
- [math]\displaystyle{ f(y) = \exp\left(\frac{y\mu - \frac{\mu^2}{2}}{\sigma^2} - \frac{y^2}{2\sigma^2} - \frac{1}{2}\ln{2\pi\sigma^2}\right) }[/math]
where
- [math]\displaystyle{ \theta = \mu, }[/math]
- [math]\displaystyle{ b(\theta) = \frac{\mu^2}{2}, }[/math]
- [math]\displaystyle{ \phi=\sigma^2, }[/math]
- [math]\displaystyle{ c(y,\phi)=- \frac{y^2}{2\sigma^2} - \frac{1}{2}\ln{2\pi\sigma^2} }[/math]
To calculate the variance function [math]\displaystyle{ V(\mu) }[/math], we first express [math]\displaystyle{ \theta }[/math] as a function of [math]\displaystyle{ \mu }[/math]. Then we transform [math]\displaystyle{ V(\theta) }[/math] into a function of [math]\displaystyle{ \mu }[/math]
- [math]\displaystyle{ \theta=\mu }[/math]
- [math]\displaystyle{ b'(\theta) = \theta= \operatorname{E}[y] = \mu }[/math]
- [math]\displaystyle{ V(\theta) = b''(\theta) = 1 }[/math]
Therefore, the variance function is constant.
Example – Bernoulli
Let [math]\displaystyle{ y \sim \text{Bernoulli}(p) }[/math], then we express the density of the Bernoulli distribution in exponential family form,
- [math]\displaystyle{ f(y) = \exp\left(y\ln\frac{p}{1-p} + \ln(1-p)\right) }[/math]
- [math]\displaystyle{ \theta = \ln\frac{p}{1-p} = }[/math] logit(p), which gives us [math]\displaystyle{ p = \frac{e^\theta}{1+e^\theta} = }[/math] expit[math]\displaystyle{ (\theta) }[/math]
- [math]\displaystyle{ b(\theta) = \ln(1+e^\theta) }[/math] and
- [math]\displaystyle{ b'(\theta) = \frac{e^\theta}{1+e^\theta} = }[/math] expit[math]\displaystyle{ (\theta) = p = \mu }[/math]
- [math]\displaystyle{ b''(\theta) =\frac{e^\theta}{1+e^\theta} - \left(\frac{e^\theta}{1+e^\theta}\right)^2 }[/math]
This give us
- [math]\displaystyle{ V(\mu) = \mu(1-\mu) }[/math]
Example – Poisson
Let [math]\displaystyle{ y \sim \text{Poisson}(\lambda) }[/math], then we express the density of the Poisson distribution in exponential family form,
- [math]\displaystyle{ f(y) = \exp(y\ln\lambda -\ln\lambda) }[/math]
- [math]\displaystyle{ \theta = \ln\lambda = }[/math] which gives us [math]\displaystyle{ \lambda = e^\theta }[/math]
- [math]\displaystyle{ b(\theta) = e^\theta }[/math] and
- [math]\displaystyle{ b'(\theta) = e^\theta = \lambda = \mu }[/math]
- [math]\displaystyle{ b''(\theta) =e^\theta = \mu }[/math]
This give us
- [math]\displaystyle{ V(\mu) = \mu }[/math]
Here we see the central property of Poisson data, that the variance is equal to the mean.
Example – Gamma
The Gamma distribution and density function can be expressed under different parametrizations. We will use the form of the gamma with parameters [math]\displaystyle{ (\mu,\nu) }[/math]
- [math]\displaystyle{ f_{\mu,\nu}(y) = \frac{1}{\Gamma(\nu)y}\left(\frac{\nu y}{\mu}\right)^\nu e^{-\frac{\nu y}{\mu}} }[/math]
Then in exponential family form we have
- [math]\displaystyle{ f_{\mu,\nu}(y) = \exp\left(\frac{-\frac{1}{\mu}y+\ln(\frac{1}{\mu})}{\frac{1}{\nu}}+ \ln\left(\frac{\nu^\nu y^{\nu-1}}{\Gamma(\nu)}\right)\right) }[/math]
- [math]\displaystyle{ \theta = \frac{-1}{\mu} \rightarrow \mu = \frac{-1}{\theta} }[/math]
- [math]\displaystyle{ \phi = \frac{1}{\nu} }[/math]
- [math]\displaystyle{ b(\theta) = -\ln(-\theta) }[/math]
- [math]\displaystyle{ b'(\theta) = \frac{-1}{\theta} = \frac{-1}{\frac{-1}{\mu}} = \mu }[/math]
- [math]\displaystyle{ b''(\theta) = \frac{1}{\theta^2} = \mu^2 }[/math]
And we have [math]\displaystyle{ V(\mu) = \mu^2 }[/math]
Application – weighted least squares
A very important application of the variance function is its use in parameter estimation and inference when the response variable is of the required exponential family form as well as in some cases when it is not (which we will discuss in quasi-likelihood). Weighted least squares (WLS) is a special case of generalized least squares. Each term in the WLS criterion includes a weight that determines that the influence each observation has on the final parameter estimates. As in regular least squares, the goal is to estimate the unknown parameters in the regression function by finding values for parameter estimates that minimize the sum of the squared deviations between the observed responses and the functional portion of the model.
While WLS assumes independence of observations it does not assume equal variance and is therefore a solution for parameter estimation in the presence of heteroscedasticity. The Gauss–Markov theorem and Aitken demonstrate that the best linear unbiased estimator (BLUE), the unbiased estimator with minimum variance, has each weight equal to the reciprocal of the variance of the measurement.
In the GLM framework, our goal is to estimate parameters [math]\displaystyle{ \beta }[/math], where [math]\displaystyle{ Z = g(E[y\mid X]) = X\beta }[/math]. Therefore, we would like to minimize [math]\displaystyle{ (Z-XB)^{T}W(Z-XB) }[/math] and if we define the weight matrix W as
- [math]\displaystyle{ \underbrace{W}_{n \times n} = \begin{bmatrix} \frac{1}{\phi V(\mu_1)g'(\mu_1)^2} &0&\cdots&0&0 \\ 0&\frac{1}{\phi V(\mu_2)g'(\mu_2)^2}&0&\cdots&0 \\ \vdots&\vdots&\vdots&\vdots&0\\ \vdots&\vdots&\vdots&\vdots&0\\ 0 &\cdots&\cdots&0&\frac{1}{\phi V(\mu_n)g'(\mu_n)^2} \end{bmatrix}, }[/math]
where [math]\displaystyle{ \phi,V(\mu),g(\mu) }[/math] are defined in the previous section, it allows for iteratively reweighted least squares (IRLS) estimation of the parameters. See the section on iteratively reweighted least squares for more derivation and information.
Also, important to note is that when the weight matrix is of the form described here, minimizing the expression [math]\displaystyle{ (Z-XB)^{T}W(Z-XB) }[/math] also minimizes the Pearson distance. See Distance correlation for more.
The matrix W falls right out of the estimating equations for estimation of [math]\displaystyle{ \beta }[/math]. Maximum likelihood estimation for each parameter [math]\displaystyle{ \beta_r, 1\leq r \leq p }[/math], requires
- [math]\displaystyle{ \sum_{i=1}^n \frac{\partial l_i}{\partial \beta_r} = 0 }[/math], where [math]\displaystyle{ \operatorname{l}(\theta,y,\phi)=\log(\operatorname{f}(y,\theta,\phi)) = \frac{y\theta - b(\theta)}{\phi} - c(y,\phi) }[/math] is the log-likelihood.
Looking at a single observation we have,
- [math]\displaystyle{ \frac{\partial l}{\partial \beta_r} = \frac{\partial l}{\partial \theta} \frac{\partial \theta}{\partial \mu}\frac{\partial \mu}{\partial \eta}\frac{\partial \eta}{\partial \beta_r} }[/math]
- [math]\displaystyle{ \frac{\partial \eta}{\partial \beta_r} = x_r }[/math]
- [math]\displaystyle{ \frac{\partial l}{\partial \theta} = \frac{y-b'(\theta)}{\phi}=\frac{y-\mu}{\phi} }[/math]
- [math]\displaystyle{ \frac{\partial \theta}{\partial \mu} = \frac{\partial b'^{-1}(\mu)}{\mu} = \frac{1}{b''(b'(\mu))} = \frac{1}{V(\mu)} }[/math]
This gives us
- [math]\displaystyle{ \frac{\partial l}{\partial \beta_r} =\frac{y-\mu}{\phi V(\mu)} \frac{\partial \mu}{\partial \eta}x_r }[/math], and noting that
- [math]\displaystyle{ \frac{\partial \eta}{\partial \mu} = g'(\mu) }[/math] we have that
- [math]\displaystyle{ \frac{\partial l}{\partial \beta_r} = (y-\mu )W\frac{\partial \eta}{\partial \mu}x_r }[/math]
The Hessian matrix is determined in a similar manner and can be shown to be,
- [math]\displaystyle{ H = X^T(y-\mu)\left[\frac{\partial}{\beta_s}W\frac{\partial}{\beta_r}\right] - X^TWX }[/math]
Noticing that the Fisher Information (FI),
- [math]\displaystyle{ \text{FI} = -E[H] = X^TWX }[/math], allows for asymptotic approximation of [math]\displaystyle{ \hat{\beta} }[/math]
- [math]\displaystyle{ \hat{\beta} \sim N_p(\beta,(X^TWX)^{-1}) }[/math], and hence inference can be performed.
Application – quasi-likelihood
Because most features of GLMs only depend on the first two moments of the distribution, rather than the entire distribution, the quasi-likelihood can be developed by just specifying a link function and a variance function. That is, we need to specify
- the link function, [math]\displaystyle{ E[y] = \mu = g^{-1}(\eta) }[/math]
- the variance function, [math]\displaystyle{ V(\mu) }[/math], where the [math]\displaystyle{ \operatorname{Var}_\theta(y) = \sigma^2 V(\mu) }[/math]
With a specified variance function and link function we can develop, as alternatives to the log-likelihood function, the score function, and the Fisher information, a quasi-likelihood, a quasi-score, and the quasi-information. This allows for full inference of [math]\displaystyle{ \beta }[/math].
Quasi-likelihood (QL)
Though called a quasi-likelihood, this is in fact a quasi-log-likelihood. The QL for one observation is
- [math]\displaystyle{ Q_i(\mu_i,y_i) = \int_{y_i}^{\mu_i} \frac{y_i-t}{\sigma^2V(t)} \, dt }[/math]
And therefore the QL for all n observations is
- [math]\displaystyle{ Q(\mu,y) = \sum_{i=1}^n Q_i(\mu_i,y_i) = \sum_{i=1}^n \int_{y_i}^{\mu_i} \frac{y-t}{\sigma^2V(t)} \, dt }[/math]
From the QL we have the quasi-score
Quasi-score (QS)
Recall the score function, U, for data with log-likelihood [math]\displaystyle{ \operatorname{l}(\mu\mid y) }[/math] is
- [math]\displaystyle{ U = \frac{\partial l }{d\mu}. }[/math]
We obtain the quasi-score in an identical manner,
- [math]\displaystyle{ U = \frac{y-\mu}{\sigma^2V(\mu)} }[/math]
Noting that, for one observation the score is
- [math]\displaystyle{ \frac{\partial Q}{\partial \mu} = \frac{y-\mu}{\sigma^2V(\mu)} }[/math]
The first two Bartlett equations are satisfied for the quasi-score, namely
- [math]\displaystyle{ E[U] = 0 }[/math]
and
- [math]\displaystyle{ \operatorname{Cov}(U) + E\left[\frac{\partial U}{\partial \mu}\right] = 0. }[/math]
In addition, the quasi-score is linear in y.
Ultimately the goal is to find information about the parameters of interest [math]\displaystyle{ \beta }[/math]. Both the QS and the QL are actually functions of [math]\displaystyle{ \beta }[/math]. Recall, [math]\displaystyle{ \mu = g^{-1}(\eta) }[/math], and [math]\displaystyle{ \eta = X\beta }[/math], therefore,
- [math]\displaystyle{ \mu = g^{-1}(X\beta). }[/math]
Quasi-information (QI)
The quasi-information, is similar to the Fisher information,
- [math]\displaystyle{ i_b = -\operatorname{E}\left[\frac{\partial U}{\partial \beta}\right] }[/math]
QL, QS, QI as functions of [math]\displaystyle{ \beta }[/math]
The QL, QS and QI all provide the building blocks for inference about the parameters of interest and therefore it is important to express the QL, QS and QI all as functions of [math]\displaystyle{ \beta }[/math].
Recalling again that [math]\displaystyle{ \mu = g^{-1}(X\beta) }[/math], we derive the expressions for QL, QS and QI parametrized under [math]\displaystyle{ \beta }[/math].
Quasi-likelihood in [math]\displaystyle{ \beta }[/math],
- [math]\displaystyle{ Q(\beta,y) = \int_y^{\mu(\beta)} \frac{y-t}{\sigma^2V(t)} \, dt }[/math]
The QS as a function of [math]\displaystyle{ \beta }[/math] is therefore
- [math]\displaystyle{ U_j(\beta_j) = \frac{\partial}{\partial \beta_j} Q(\beta,y) = \sum_{i=1}^n \frac{\partial \mu_i}{\partial\beta_j} \frac{y_i-\mu_i(\beta_j)}{\sigma^2V(\mu_i)} }[/math]
- [math]\displaystyle{ U(\beta) = \begin{bmatrix} U_1(\beta)\\ U_2(\beta)\\ \vdots\\ \vdots\\ U_p(\beta) \end{bmatrix} = D^TV^{-1}\frac{(y-\mu)}{\sigma^2} }[/math]
Where,
- [math]\displaystyle{ \underbrace{D}_{n \times p} = \begin{bmatrix} \frac{\partial \mu_1}{\partial \beta_1} &\cdots&\cdots&\frac{\partial \mu_1}{\partial \beta_p} \\ \frac{\partial \mu_2}{\partial \beta_1} &\cdots&\cdots&\frac{\partial \mu_2}{\partial \beta_p} \\ \vdots\\ \vdots\\ \frac{\partial \mu_m}{\partial \beta_1} &\cdots&\cdots&\frac{\partial \mu_m}{\partial \beta_p} \end{bmatrix} \underbrace{V}_{n\times n} = \operatorname{diag}(V(\mu_1),V(\mu_2),\ldots,\ldots,V(\mu_n)) }[/math]
The quasi-information matrix in [math]\displaystyle{ \beta }[/math] is,
- [math]\displaystyle{ i_b = -\frac{\partial U}{\partial \beta} = \operatorname{Cov}(U(\beta)) = \frac{D^TV^{-1}D}{\sigma^2} }[/math]
Obtaining the score function and the information of [math]\displaystyle{ \beta }[/math] allows for parameter estimation and inference in a similar manner as described in Application – weighted least squares.
Non-parametric regression analysis
Non-parametric estimation of the variance function and its importance, has been discussed widely in the literature[5][6][7] In non-parametric regression analysis, the goal is to express the expected value of your response variable(y) as a function of your predictors (X). That is we are looking to estimate a mean function, [math]\displaystyle{ g(x) = \operatorname{E}[y\mid X=x] }[/math] without assuming a parametric form. There are many forms of non-parametric smoothing methods to help estimate the function [math]\displaystyle{ g(x) }[/math]. An interesting approach is to also look at a non-parametric variance function, [math]\displaystyle{ g_v(x) = \operatorname{Var}(Y\mid X=x) }[/math]. A non-parametric variance function allows one to look at the mean function as it relates to the variance function and notice patterns in the data.
- [math]\displaystyle{ g_v(x) = \operatorname{Var}(Y\mid X=x) =\operatorname{E}[y^2\mid X=x] - \left[\operatorname{E}[y\mid X=x]\right]^2 }[/math]
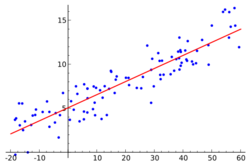
An example is detailed in the pictures to the right. The goal of the project was to determine (among other things) whether or not the predictor, number of years in the major leagues (baseball), had an effect on the response, salary, a player made. An initial scatter plot of the data indicates that there is heteroscedasticity in the data as the variance is not constant at each level of the predictor. Because we can visually detect the non-constant variance, it useful now to plot [math]\displaystyle{ g_v(x) = \operatorname{Var}(Y\mid X=x) =\operatorname{E}[y^2\mid X=x] - \left[\operatorname{E}[y\mid X=x]\right]^2 }[/math], and look to see if the shape is indicative of any known distribution. One can estimate [math]\displaystyle{ \operatorname{E}[y^2\mid X=x] }[/math] and [math]\displaystyle{ \left[\operatorname{E}[y\mid X=x]\right]^2 }[/math] using a general smoothing method. The plot of the non-parametric smoothed variance function can give the researcher an idea of the relationship between the variance and the mean. The picture to the right indicates a quadratic relationship between the mean and the variance. As we saw above, the Gamma variance function is quadratic in the mean.
Notes
- ↑ 1.0 1.1 Muller and Zhao (1995). "On a semi parametric variance function model and a test for heteroscedasticity". The Annals of Statistics 23 (3): 946–967. doi:10.1214/aos/1176324630.
- ↑ Muller, Stadtmuller and Yao (2006). "Functional Variance Processes". Journal of the American Statistical Association 101 (475): 1007–1018. doi:10.1198/016214506000000186.
- ↑ Wedderburn, R.W.M. (1974). "Quasi-likelihood functions, generalized linear models, and the Gauss–Newton Method". Biometrika 61 (3): 439–447. doi:10.1093/biomet/61.3.439.
- ↑ McCullagh, Peter; Nelder, John (1989). Generalized Linear Models (second ed.). London: Chapman and Hall. ISBN 0-412-31760-5.
- ↑ Muller and StadtMuller (1987). "Estimation of Heteroscedasticity in Regression Analysis". The Annals of Statistics 15 (2): 610–625. doi:10.1214/aos/1176350364.
- ↑ Cai and Wang, T.; Wang, Lie (2008). "Adaptive Variance Function Estimation in Heteroscedastic Nonparametric Regression". The Annals of Statistics 36 (5): 2025–2054. doi:10.1214/07-AOS509. Bibcode: 2008arXiv0810.4780C.
- ↑ Rice and Silverman (1991). "Estimating the Mean and Covariance structure nonparametrically when the data are curves". Journal of the Royal Statistical Society 53 (1): 233–243.
References
- McCullagh, Peter; Nelder, John (1989). Generalized Linear Models (second ed.). London: Chapman and Hall. ISBN 0-412-31760-5.
- Henrik Madsen and Poul Thyregod (2011). Introduction to General and Generalized Linear Models. Chapman & Hall/CRC. ISBN 978-1-4200-9155-7.
External links
 |



