Biology:P2RX4
 Generic protein structure example |
P2X purinoceptor 4 is a protein that in humans is encoded by the P2RX4 gene. P2X purinoceptor 4 is a member of the P2X receptor family.[1][2][3] P2X receptors are trimeric protein complexes that can be homomeric or heteromeric. These receptors are ligand-gated cation channels that open in response to ATP binding.[4] Each receptor subtype, determined by the subunit composition, varies in its affinity to ATP and desensitization kinetics.
The P2X4 receptor is the homotrimer composed of three P2X4 monomers.[1] They are nonselective cation channels with high calcium permeability, leading to the depolarization of the cell membrane and the activation of various Ca2+-sensitive intracellular processes.[5][6][7] The P2X4 receptor is uniquely expressed on lysosomal compartments as well as the cell surface.[8]
The receptor is found in the central and peripheral nervous systems, in the epithelia of ducted glands and airways, in the smooth muscle of the bladder, gastrointestinal tract, uterus, and arteries, in uterine endometrium, and in fat cells.[9] P2X4 receptors have been implicated in the regulation of cardiac function, ATP-mediated cell death, synaptic strengthening, and activating of the inflammasome in response to injury.[8][10][11][12][13][14]
Structure

P2X receptors are composed of three subunits that can be homomeric or heteromeric by nature. In mammals, there are seven different subunits, each encoded in a different gene (P2RX1-P2RX7).[1] Each subunit has two transmembrane alpha helices (TM1 and TM2) linked by a large extracellular loop.[1][8][15] Analysis of x-ray crystallographic structures revealed a 'dolphin-like' tertiary structure, where the 'tail' is embedded in the phospholipid bilayer and the upper and lower ectodomains form the 'head' and 'body' respectively.[8][15][16] Adjacent interfaces of the subunits form a deep binding pocket for ATP.[8][15] ATP binding to these orthosteric sites causes a shift in conformation opening the channel pore.
The P2X4 subunits can form homomeric or heteromeric receptors.[17] In 2009, the first purinergic receptor crystallized was the closed state homomeric zebrafish P2X4 receptor.[18][15] Although truncated at its N- and C- termini, this crystal structure resolved and confirmed that these proteins were indeed trimers with an ectodomain rich with disulfide bonds.[1][8]
Gating mechanism
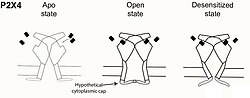
P2X receptors have three confirmed conformational states: ATP-unbound closed, ATP-bound open, and ATP-bound desensitized.[8][15] Imaging of the human P2X3 and rat P2X7 receptors has revealed structural similarities and differences in their cytoplasmic domains. In the ATP-bound state, both receptor types form beta sheet structures from N- and C- termini of adjacent subunits.[8][15] These newly folded secondary structures come together to form a 'cytoplasmic cap' that helps stabilize the open pore. Crystal structures of the desensitized receptor no longer exhibit the cytoplasmic cap.[8][15]
Desensitization
Electrophysiology studies have revealed differences in the rates of receptor desensitization between different P2X subtypes.[1][8] Homotrimers P2X1 and P2X3 are the fastest, with desensitization observed milliseconds after activation, while P2X2 and P2X4 receptors are on the timescale of seconds. Notably, the P2X7 receptor uniquely does not undergo desensitization.[8] Mutational studies working with the rat P2X2 and P2X3 receptors have identified three residues in the N-terminus that majorly contribute to these differences. By changing the amino acids in the P2X3 to match the analogous P2X2, the desensitization rate slowed down. Conversely, changing residues of P2X2 to match P2X3 increased the desensitization rate.[15] In combination with the open state crystal structures, it was hypothesized that the cytoplasmic cap was stabilizing the open pore conformation.[8][15]
Additionally, structural analysis of the open P2X3 receptor revealed transient changes in TM2, the transmembrane alpha helix lining the pore. While in the open state conformation, a small mid-region of TM2 develops into a 310-helix.[8][15] This helical structure disappears with desensitization and instead TM2 reforms as a complete alpha helix repositioned closer to the extracellular side.[8]
The helical recoil model uses the observed structural changes in TM2 and the transient formation of the cytoplasmic cap to describe a possible mechanism for the desensitization of P2X receptors. In this model, it is theorized that the cytoplasmic cap fixes the intracellular end of the TM2 helix while stretching its extracellular end to allow ion influx.[15] This would induce the observed 310-helix. The cap then disassembles and releases its hold on TM2 causing the helix to recoil towards the outer leaflet of the membrane.[8][15]
In support of this theory, the P2X7 uniquely has a large cytoplasmic domain with palmitoylated C-cysteine anchor sites.[1][8][15] These sites further stabilize its cytoplasmic cap by anchoring the domain into the surrounding inner leaflet. Mutations of the associated palmitoylation site residues cause observed atypical desensitization of the receptor.[8]
Receptor trafficking
P2X4 receptors are functionally expressed on both the cell surface and in lysosomes.[16] Although preferentially localized and stored in lysosomes, P2X4 receptors are brought to the cell surface in response to extracellular signals.[19] These signals include IFN-γ, CCL21, CCL2.[20][21][22] Fibronectin is also involved in upregulation of P2X4 receptors through interactions with integrins that lead to the activation of SRC-family kinase member, Lyn.[23] Lyn then activates PI3K-AKT and MEK-ERK signaling pathways to stimulate receptor trafficking.[24] Internalization of P2X4 receptors is clathrin- and dynamin-dependent endocytosis.[25]
Pharmacology
Agonists
P2X4 receptors respond to ATP, but not αβmeATP. These receptors are also potentiated by ivermectin, cibacron blue, and zinc.[4]
Antagonists
The main pharmacological distinction between the members of the purinoceptor family is the relative sensitivity to the antagonists suramin and pyridoxalphosphate-6-azophenyl-2',4'-disulphonic acid (PPADS). The product of this gene has the lowest sensitivity for these antagonists[4]
Neuropathic pain
The P2X4 receptor has been linked to neuropathic pain mediated by microglia in vitro and in vivo.[26][27] P2X4 receptors are upregulated following injury.[28] This upregulation allows for increased activation of p38 mitogen-activated protein kinases, thereby increasing the release of brain-derived neurotrophic factor (BDNF) from microglia.[29] BDNF released from microglia induces neuronal hyperexcitability through interaction with the TrkB receptor.[30] More importantly, recent work shows that P2X4 receptor activation is not only necessary for neuropathic pain, but it is also sufficient to cause neuropathic pain.[31]
See also
- P2X receptor
References
- ↑ 1.0 1.1 1.2 1.3 1.4 1.5 1.6 "P2X4: A fast and sensitive purinergic receptor". Biomedical Journal 40 (5): 245–256. October 2017. doi:10.1016/j.bj.2017.06.010. PMID 29179879.
- ↑ "Characterization of recombinant human P2X4 receptor reveals pharmacological differences to the rat homologue". Molecular Pharmacology 51 (1): 109–118. January 1997. doi:10.1124/mol.51.1.109. PMID 9016352.
- ↑ "Entrez Gene: P2RX4 purinergic receptor P2X, ligand-gated ion channel, 4". https://www.ncbi.nlm.nih.gov/sites/entrez?Db=gene&Cmd=ShowDetailView&TermToSearch=5025.
- ↑ 4.0 4.1 4.2 "Molecular physiology of P2X receptors". Physiological Reviews 82 (4): 1013–1067. October 2002. doi:10.1152/physrev.00015.2002. PMID 12270951.
- ↑ "Molecular physiology of P2X receptors". Physiological Reviews 82 (4): 1013–1067. October 2002. doi:10.1152/physrev.00015.2002. PMID 12270951.
- ↑ "Action potential-independent release of glutamate by Ca2+ entry through presynaptic P2X receptors elicits postsynaptic firing in the brainstem autonomic network". The Journal of Neuroscience 24 (12): 3125–3135. March 2004. doi:10.1523/JNEUROSCI.0090-04.2004. PMID 15044552.
- ↑ "Characterization of calcium signaling by purinergic receptor-channels expressed in excitable cells". Molecular Pharmacology 58 (5): 936–945. November 2000. doi:10.1124/mol.58.5.936. PMID 11040040.
- ↑ 8.00 8.01 8.02 8.03 8.04 8.05 8.06 8.07 8.08 8.09 8.10 8.11 8.12 8.13 8.14 8.15 8.16 "Structural and Functional Features of the P2X4 Receptor: An Immunological Perspective". Frontiers in Immunology 12: 645834. 2021-03-25. doi:10.3389/fimmu.2021.645834. PMID 33897694.
- ↑ "Tissue distribution of P2X4 receptors studied with an ectodomain antibody". Cell and Tissue Research 313 (2): 159–165. August 2003. doi:10.1007/s00441-003-0758-5. PMID 12845522.
- ↑ "Involvement of P2X4 receptor in P2X7 receptor-dependent cell death of mouse macrophages". Biochemical and Biophysical Research Communications 419 (2): 374–380. March 2012. doi:10.1016/j.bbrc.2012.01.156. PMID 22349510.
- ↑ "Multiple P2X receptors are involved in the modulation of apoptosis in human mesangial cells: evidence for a role of P2X4". American Journal of Physiology. Renal Physiology 292 (5): F1537–F1547. May 2007. doi:10.1152/ajprenal.00440.2006. PMID 17264311.
- ↑ "Extracellular ATP-stimulated current in wild-type and P2X4 receptor transgenic mouse ventricular myocytes: implications for a cardiac physiologic role of P2X4 receptors". FASEB Journal 20 (2): 277–284. February 2006. doi:10.1096/fj.05-4749com. PMID 16449800.
- ↑ "Role of P2X4 receptors in synaptic strengthening in mouse CA1 hippocampal neurons". The European Journal of Neuroscience 34 (2): 213–220. July 2011. doi:10.1111/j.1460-9568.2011.07763.x. PMID 21749490.
- ↑ "P2X4 receptors influence inflammasome activation after spinal cord injury". The Journal of Neuroscience 32 (9): 3058–3066. February 2012. doi:10.1523/JNEUROSCI.4930-11.2012. PMID 22378878.
- ↑ 15.00 15.01 15.02 15.03 15.04 15.05 15.06 15.07 15.08 15.09 15.10 15.11 15.12 "How Structural Biology Has Directly Impacted Our Understanding of P2X Receptor Function and Gating". The P2X7 Receptor. Methods in Molecular Biology. 2510. New York, NY: Springer US. 2022. pp. 1–29. doi:10.1007/978-1-0716-2384-8_1. ISBN 978-1-0716-2384-8.
- ↑ 16.0 16.1 "The P2X4 Receptor: Cellular and Molecular Characteristics of a Promising Neuroinflammatory Target". International Journal of Molecular Sciences 23 (10): 5739. May 2022. doi:10.3390/ijms23105739. PMID 35628550.
- ↑ "Molecular and functional properties of P2X receptors--recent progress and persisting challenges". Purinergic Signalling 8 (3): 375–417. September 2012. doi:10.1007/s11302-012-9314-7. PMID 22547202.
- ↑ "Crystal structure of the ATP-gated P2X(4) ion channel in the closed state". Nature 460 (7255): 592–598. July 2009. doi:10.1038/nature08198. PMID 19641588. Bibcode: 2009Natur.460..592K.
- ↑ "Regulation of P2X4 receptors by lysosomal targeting, glycan protection and exocytosis". Journal of Cell Science 120 (Pt 21): 3838–3849. November 2007. doi:10.1242/jcs.010348. PMID 17940064.
- ↑ "IFN-gamma receptor signaling mediates spinal microglia activation driving neuropathic pain". Proceedings of the National Academy of Sciences of the United States of America 106 (19): 8032–8037. May 2009. doi:10.1073/pnas.0810420106. PMID 19380717. Bibcode: 2009PNAS..106.8032T.
- ↑ "Neuronal CCL21 up-regulates microglia P2X4 expression and initiates neuropathic pain development". The EMBO Journal 30 (9): 1864–1873. May 2011. doi:10.1038/emboj.2011.89. PMID 21441897.
- ↑ "CCL2 promotes P2X4 receptor trafficking to the cell surface of microglia". Purinergic Signalling 8 (2): 301–310. June 2012. doi:10.1007/s11302-011-9288-x. PMID 22222817.
- ↑ "Lyn tyrosine kinase is required for P2X(4) receptor upregulation and neuropathic pain after peripheral nerve injury". Glia 56 (1): 50–58. January 2008. doi:10.1002/glia.20591. PMID 17918263.
- ↑ "Mechanisms underlying fibronectin-induced up-regulation of P2X4R expression in microglia: distinct roles of PI3K-Akt and MEK-ERK signalling pathways". Journal of Cellular and Molecular Medicine 13 (9B): 3251–3259. September 2009. doi:10.1111/j.1582-4934.2009.00719.x. PMID 19298529.
- ↑ "Identification of a non-canonical tyrosine-based endocytic motif in an ionotropic receptor". The Journal of Biological Chemistry 277 (38): 35378–35385. September 2002. doi:10.1074/jbc.M204844200. PMID 12105201.
- ↑ "P2X4 receptors mediate PGE2 release by tissue-resident macrophages and initiate inflammatory pain". The EMBO Journal 29 (14): 2290–2300. July 2010. doi:10.1038/emboj.2010.126. PMID 20562826.
- ↑ "Behavioral phenotypes of mice lacking purinergic P2X4 receptors in acute and chronic pain assays". Molecular Pain 5: 28. June 2009. doi:10.1186/1744-8069-5-28. PMID 19515262.
- ↑ "Up-regulation of P2X4 receptors in spinal microglia after peripheral nerve injury mediates BDNF release and neuropathic pain". The Journal of Neuroscience 28 (44): 11263–11268. October 2008. doi:10.1523/JNEUROSCI.2308-08.2008. PMID 18971468.
- ↑ "P2X4-receptor-mediated synthesis and release of brain-derived neurotrophic factor in microglia is dependent on calcium and p38-mitogen-activated protein kinase activation". The Journal of Neuroscience 29 (11): 3518–3528. March 2009. doi:10.1523/JNEUROSCI.5714-08.2009. PMID 19295157.
- ↑ "BDNF from microglia causes the shift in neuronal anion gradient underlying neuropathic pain". Nature 438 (7070): 1017–1021. December 2005. doi:10.1038/nature04223. PMID 16355225. Bibcode: 2005Natur.438.1017C.
- ↑ "P2X4 receptors induced in spinal microglia gate tactile allodynia after nerve injury". Nature 424 (6950): 778–783. August 2003. doi:10.1038/nature01786. PMID 12917686. Bibcode: 2003Natur.424..778T.
Further reading
- "Molecular physiology of P2X receptors". Physiological Reviews 82 (4): 1013–1067. October 2002. doi:10.1152/physrev.00015.2002. PMID 12270951.
- "Oligo-capping: a simple method to replace the cap structure of eukaryotic mRNAs with oligoribonucleotides". Gene 138 (1–2): 171–174. January 1994. doi:10.1016/0378-1119(94)90802-8. PMID 8125298.
- "Molecular characterization and pharmacological properties of the human P2X3 purinoceptor". Brain Research. Molecular Brain Research 47 (1–2): 59–66. July 1997. doi:10.1016/S0169-328X(97)00036-3. PMID 9221902.
- "Construction and characterization of a full length-enriched and a 5'-end-enriched cDNA library". Gene 200 (1–2): 149–156. October 1997. doi:10.1016/S0378-1119(97)00411-3. PMID 9373149.
- "Sp1-mediated downregulation of P2X4 receptor gene transcription in endothelial cells exposed to shear stress". American Journal of Physiology. Heart and Circulatory Physiology 280 (5): H2214–H2221. May 2001. doi:10.1152/ajpheart.2001.280.5.H2214. PMID 11299224.
- "P2X4 and P2X6 receptors associate with VE-cadherin in human endothelial cells". Cellular and Molecular Life Sciences 59 (5): 870–881. May 2002. doi:10.1007/s00018-002-8474-y. PMID 12088286.
- "Endogenously released ATP mediates shear stress-induced Ca2+ influx into pulmonary artery endothelial cells". American Journal of Physiology. Heart and Circulatory Physiology 285 (2): H793–H803. August 2003. doi:10.1152/ajpheart.01155.2002. PMID 12714321.
- "Enhanced expression of the P2X4 receptor in Duchenne muscular dystrophy correlates with macrophage invasion". Neurobiology of Disease 15 (2): 212–220. March 2004. doi:10.1016/j.nbd.2003.10.014. PMID 15006691.
- "A beneficial role of cardiac P2X4 receptors in heart failure: rescue of the calsequestrin overexpression model of cardiomyopathy". American Journal of Physiology. Heart and Circulatory Physiology 287 (3): H1096–H1103. September 2004. doi:10.1152/ajpheart.00079.2004. PMID 15130891.
- "cAMP potentiates ATP-evoked calcium signaling in human parotid acinar cells". The Journal of Biological Chemistry 279 (38): 39485–39494. September 2004. doi:10.1074/jbc.M406201200. PMID 15262999.
- "A C-terminal lysine that controls human P2X4 receptor desensitization". The Journal of Biological Chemistry 281 (22): 15044–15049. June 2006. doi:10.1074/jbc.M600442200. PMID 16533808.
- "Identification of P2X4 receptor-specific residues contributing to the ivermectin effects on channel deactivation". Biochemical and Biophysical Research Communications 349 (2): 619–625. October 2006. doi:10.1016/j.bbrc.2006.08.084. PMID 16949036.
- "Multiple P2X receptors are involved in the modulation of apoptosis in human mesangial cells: evidence for a role of P2X4". American Journal of Physiology. Renal Physiology 292 (5): F1537–F1547. May 2007. doi:10.1152/ajprenal.00440.2006. PMID 17264311.
External links
- P2RX4+protein,+human at the US National Library of Medicine Medical Subject Headings (MeSH)
This article incorporates text from the United States National Library of Medicine, which is in the public domain.
 |
