Biology:Bestrophin 1
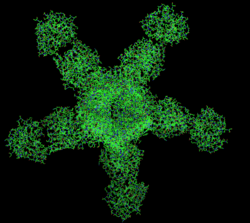
 Generic protein structure example |
Bestrophin-1 (Best1) is a protein that, in humans, is encoded by the BEST1 gene (RPD ID - 5T5N/4RDQ).[1]
The bestrophin family of proteins comprises four evolutionary related genes (BEST1, BEST2, BEST3, and BEST4) that code for integral membrane proteins.[2] This family was first identified in humans by linking a BEST1 mutation with Best vitelliform macular dystrophy (BVMD).[3] Mutations in the BEST1 gene have been identified as the primary cause for at least five different degenerative retinal diseases.[3]
The bestrophins are an ancient family of structurally conserved proteins that have been identified in nearly every organism studied from bacteria to humans. In humans, they function as calcium-activated anion channels, each of which has a unique tissue distribution throughout the body. Specifically, the BEST1 gene on chromosome 11q13 encodes the Bestrophin-1 protein in humans whose expression is highest in the retina.[3]
Structure
Gene
The bestrophin genes share a conserved gene structure, with almost identical sizes of the 8 RFP-TM domain-encoding exons and highly conserved exon-intron boundaries. Each of the four bestrophin genes has a unique 3-prime end of variable length.[1]
BEST1 has been shown by two independent studies to be regulated by Microphthalmia-associated transcription factor.[4][5]
Protein
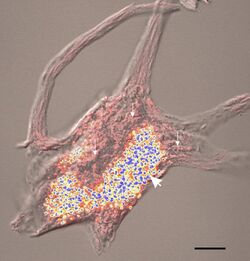
Bestrophin-1 is an integral membrane protein found primarily in the retinal pigment epithelium (RPE) of the eye.[6] Within the RPE layer, it is mainly located on the basolateral plasma membrane. Protein crystallization structures indicate this protein's primary ion channel function as well as its calcium regulatory capabilities.[6][3] Bestrophin-1 consists of 585 amino acids and both N- and the C-termini are located within the cell.
The structure of Best1 consists of five identical subunits that each span the membrane four times and form a continuous, funnel-shaped pore via the second transmembrane domain containing a high content of aromatic residues, including an invariant arg-phe-pro (RFP) motif.[3][7][8] The pore is lined with various nonpolar, hydrophobic amino acids. Both the structure and the composition of the pore help to ensure that only small anions are able to move completely through the channel. The channel acts as two funnels working together in tandem. It begins with a semi-selective, narrow entryway for anions, and then opens to a larger, positively charged area which then leads to a narrower pathway that further limits the size of anions passing through the pore. A calcium clasp acts as a belting mechanism around the larger, middle section of the channel. Calcium ions control the opening and closing of the channel due to conformational changes caused by calcium binding at the C-terminus directly following the last transmembrane domain.[3][8]
Tissue and subcellular distribution
The location of expression of the BEST1 gene is essential for protein functioning and mislocalization is often connected to a variety of retinal degenerative diseases. The BEST1 gene expresses the Best1 protein primarily in the cytosol of the retinal pigment epithelium. The protein is typically contained in vesicles near the cellular membrane. There is also research to support that the Best1 protein is localized and produced in the endoplasmic reticulum (intracellular organelle involved in protein and lipid synthesis). Best1 is typically expressed with other proteins also synthesized in the endoplasmic reticulum, such as calreticulin, calnexin and Stim-1. Calcium ion involvement in the countertransport of chloride ions also supports the idea that Best1 is involved in forming calcium stores within the cell.[6]
Function
Best1 primarily functions as an intracellular calcium-activated chloride channel on the cellular membrane that is not voltage-dependent.[2][6][8] More recently Best1 has been shown to act as a volume-regulating anion channel.
Diseases
Best vitelliform macular dystrophy (BVMD)
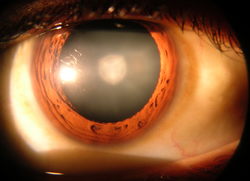
Best's vitelliform macular dystrophy (BVMD) is one of the most common Best1-associated diseases. BVMD typically becomes noticeable in children and is represented by the buildup of lipofuscin (lipid residuals) lesions in the eye.[2][6] Diagnosis normally follows an abnormal electrooculogram in which decreased activation of calcium channels in the basolateral membrane of the retinal pigment epithelium becomes apparent. A mutation in the BEST1 gene leads to a loss of channel function and eventually retinal degeneration.[6] Although BVMD is an autosomal dominant form of macular dystrophy, expressivity varies within and between affected families although the overwhelming majority of affected families come from northern European descent.[3][6] Typically, people with this condition experience five progressively worsening stages, though timing and severity varies greatly. BVMD is often caused by the single missense mutations; however, amino acid deletions have also been identified.[3] A loss of function of the Best1 chloride channel could likely explain some of the most common issues associated with BVMD: an inability to regulate intracellular ion concentrations and regulate overall cell volume.[9] To date, over 100 disease-causing mutations have been related to BVMD as well as a number of other degenerative retinal diseases.[8]
Adult-onset vitelliform macular dystrophy (AVMD)
Adult-onset vitelliform macular dystrophy (AVMD) consists of lesions similar to BVMD on the retina. However, the cause is not as definitive as BVMD. The inability to diagnosis AVMD via genetic testing makes differentiating between AVMD and pattern dystrophy difficult. It is also unknown whether there is truly a clinical difference between AVMD caused by BEST1 mutations and AVMD caused by PRPH2 mutations. AVMD usually involves less vision loss than BVMD and cases do not usually run in families.[3]
Autosomal recessive bestrophinopathy (ARB)
Autosomal recessive bestrophinopathy (ARB) was first identified in 2008. People with ARB demonstrate a decrease in vision during the first ten years of life. Parents and family members typically show no abnormalities as the disease is autosomal recessive, indicating that both alleles of the BEST1 gene must be mutated. Vitelliform lesions are often present and some cases involve cystoid macular edema. In addition, other complications have been observed. Vision decreases slowly over time, although rates of decline vary. Mutations causing ARB range from missense mutations to single base mutations in non-coding regions.[3]
Autosomal dominant vitreoretinochoroidopathy
Autosomal dominant vitreoretinochoroidopathy was first identified in 1982 and presents itself in both eyes with decreases in peripheral vision due to excessive fluid and changes in eye retinal pigmentation. Early onset cataracts are also likely.[3]
Retinitis pigmentosa (RP)
Retinitis pigmentosa was first described in relation to the BEST1 gene in 2009 and was found to be associated with four different missense mutations in the BEST1 gene in people. All affected individuals experience a diminished response to light within their retina and may have changes in pigmentation, pale optic discs, fluid accumulation and decreased visual acuity.[3]
All of the diseases above do not have any known treatments or cures. However, as of 2017, researchers are currently working on discovering treatments with stem cell transplants of the retinal pigment epithelium.[3]
References
- ↑ 1.0 1.1 "Entrez Gene: BEST1 bestrophin 1". https://www.ncbi.nlm.nih.gov/sites/entrez?Db=gene&Cmd=ShowDetailView&TermToSearch=7439.
- ↑ 2.0 2.1 2.2 "TMEM16, LRRC8A, bestrophin: chloride channels controlled by Ca(2+) and cell volume". Trends in Biochemical Sciences 40 (9): 535–43. September 2015. doi:10.1016/j.tibs.2015.07.005. PMID 26254230.
- ↑ 3.00 3.01 3.02 3.03 3.04 3.05 3.06 3.07 3.08 3.09 3.10 3.11 3.12 "Bestrophin 1 and retinal disease". Progress in Retinal and Eye Research 58: 45–69. January 2017. doi:10.1016/j.preteyeres.2017.01.006. PMID 28153808.
- ↑ "VMD2 promoter requires two proximal E-box sites for its activity in vivo and is regulated by the MITF-TFE family". The Journal of Biological Chemistry 282 (3): 1838–50. January 2007. doi:10.1074/jbc.M609517200. PMID 17085443.
- ↑ "Novel MITF targets identified using a two-step DNA microarray strategy". Pigment Cell & Melanoma Research 21 (6): 665–76. December 2008. doi:10.1111/j.1755-148X.2008.00505.x. PMID 19067971.
- ↑ 6.0 6.1 6.2 6.3 6.4 6.5 6.6 "A potential cytosolic function of bestrophin-1". Retinal Degenerative Diseases. Advances in Experimental Medicine and Biology. 723. 2012. pp. 603–10. doi:10.1007/978-1-4614-0631-0_77. ISBN 978-1-4614-0630-3.
- ↑ "Molecular physiology of bestrophins: multifunctional membrane proteins linked to best disease and other retinopathies". Physiological Reviews 88 (2): 639–72. April 2008. doi:10.1152/physrev.00022.2007. PMID 18391176.
- ↑ 8.0 8.1 8.2 8.3 "Bestrophins and retinopathies". Pflügers Archiv 460 (2): 559–69. July 2010. doi:10.1007/s00424-010-0821-5. PMID 20349192.
- ↑ "The Role of Bestrophin-1 in Intracellular Ca2+ Signaling". Retinal Degenerative Diseases: Mechanisms and Experimental Therapy. Advances in Experimental Medicine and Biology. 801. New York: Springer. 2014. pp. 113–119. doi:10.1007/978-1-4614-3209-8_15. ISBN 978-1-4614-3209-8.
Further reading
- "VMD2 mutations in vitelliform macular dystrophy (Best disease) and other maculopathies". Human Mutation 15 (4): 301–8. 2000. doi:10.1002/(SICI)1098-1004(200004)15:4<301::AID-HUMU1>3.0.CO;2-N. PMID 10737974.
- "Hereditary maculardegeneration (HMD) in 246 cases traced to one gene-source in central Sweden". Hereditas 84 (2): 163–76. February 1977. doi:10.1111/j.1601-5223.1977.tb01394.x. PMID 838599.
- "The gene for Best's macular dystrophy is located at 11q13 in a Swedish family". Clinical Genetics 42 (3): 156–9. September 1992. doi:10.1111/j.1399-0004.1992.tb03229.x. PMID 1395087.
- "A gene map of the Best's vitelliform macular dystrophy region in chromosome 11q12-q13.1". Genome Research 8 (1): 48–56. January 1998. doi:10.1101/gr.8.1.48. PMID 9445487.
- "Identification of the gene responsible for Best macular dystrophy". Nature Genetics 19 (3): 241–7. July 1998. doi:10.1038/915. PMID 9662395.
- "New gene found for inherited macular degeneration". Science 281 (5373): 31. July 1998. doi:10.1126/science.281.5373.31. PMID 9679014.
- "Mutations in a novel gene, VMD2, encoding a protein of unknown properties cause juvenile-onset vitelliform macular dystrophy (Best's disease)". Human Molecular Genetics 7 (9): 1517–25. September 1998. doi:10.1093/hmg/7.9.1517. PMID 9700209.
- "Bestrophin gene mutations in patients with Best vitelliform macular dystrophy". Genomics 58 (1): 98–101. May 1999. doi:10.1006/geno.1999.5808. PMID 10331951.
- "The mutation spectrum of the bestrophin protein--functional implications". Human Genetics 104 (5): 383–9. May 1999. doi:10.1007/s004390050972. PMID 10394929.
- "Evaluation of the Best disease gene in patients with age-related macular degeneration and other maculopathies". Human Genetics 104 (6): 449–53. June 1999. doi:10.1007/s004390050986. PMID 10453731.
- "A novel spontaneous missense mutation in VMD2 gene is a cause of a best macular dystrophy sporadic case". American Journal of Ophthalmology 129 (2): 260–2. February 2000. doi:10.1016/S0002-9394(99)00327-X. PMID 10682987.
- "Mutation analysis of 3 genes in patients with Leber congenital amaurosis". Archives of Ophthalmology 118 (4): 538–43. April 2000. doi:10.1001/archopht.118.4.538. PMID 10766140.
- "Allelic variation in the VMD2 gene in best disease and age-related macular degeneration". Investigative Ophthalmology & Visual Science 41 (6): 1291–6. May 2000. PMID 10798642.
- "Mutations in the VMD2 gene are associated with juvenile-onset vitelliform macular dystrophy (Best disease) and adult vitelliform macular dystrophy but not age-related macular degeneration". European Journal of Human Genetics 8 (4): 286–92. April 2000. doi:10.1038/sj.ejhg.5200447. PMID 10854112.
- "Bestrophin, the product of the Best vitelliform macular dystrophy gene (VMD2), localizes to the basolateral plasma membrane of the retinal pigment epithelium". Proceedings of the National Academy of Sciences of the United States of America 97 (23): 12758–63. November 2000. doi:10.1073/pnas.220402097. PMID 11050159. Bibcode: 2000PNAS...9712758M.
- "Identification of novel VMD2 gene mutations in patients with best vitelliform macular dystrophy". Human Mutation 17 (3): 235. March 2001. doi:10.1002/humu.9. PMID 11241846.
- "Best's vitelliform macular dystrophy caused by a new mutation (Val89Ala) in the VMD2 gene". Ophthalmic Genetics 22 (2): 107–15. June 2001. doi:10.1076/opge.22.2.107.2226. PMID 11449320.
- "The vitelliform macular dystrophy protein defines a new family of chloride channels". Proceedings of the National Academy of Sciences of the United States of America 99 (6): 4008–13. March 2002. doi:10.1073/pnas.052692999. PMID 11904445. Bibcode: 2002PNAS...99.4008S.
- "Dysregulation of human bestrophin-1 by ceramide-induced dephosphorylation". The Journal of Physiology 587 (Pt 18): 4379–91. September 2009. doi:10.1113/jphysiol.2009.176800. PMID 19635817.
- "Regulation of bestrophin Cl channels by calcium: role of the C terminus". The Journal of General Physiology 132 (6): 681–92. December 2008. doi:10.1085/jgp.200810056. PMID 19029375.
External links
- GeneReviews/NCBI/NIH/UW entry on Retinitis Pigmentosa Overview
- Human BEST1 genome location and BEST1 gene details page in the UCSC Genome Browser.
This article incorporates text from the United States National Library of Medicine, which is in the public domain.
 |