Biology:Nuclear receptor

In the field of molecular biology, nuclear receptors are a class of proteins responsible for sensing steroids, thyroid hormones, vitamins, and certain other molecules. These intracellular receptors work with other proteins to regulate the expression of specific genes thereby controlling the development, homeostasis, and metabolism of the organism.
Nuclear receptors bind directly to DNA regulating the expression of adjacent genes; hence these receptors are classified as transcription factors.[2][3] The regulation of gene expression by nuclear receptors often occurs in the presence of a ligand—a molecule that affects the receptor's behavior. Ligand binding to a nuclear receptor results in a conformational change activating the receptor. The result is up- or down-regulation of gene expression.
A unique property of nuclear receptors that differentiates them from other classes of receptors is their direct control of genomic DNA. Nuclear receptors play key roles in both embryonic development and adult homeostasis. As discussed below nuclear receptors are classified according to mechanism[4][5] or homology.[6][7]
Species distribution
Nuclear receptors are specific to metazoans (animals) and are not found in protists, algae, fungi, or plants.[8] Amongst the early-branching animal lineages with sequenced genomes, two have been reported from the sponge Amphimedon queenslandica, two from the comb jelly Mnemiopsis leidyi[9] four from the placozoan Trichoplax adhaerens and 17 from the cnidarian Nematostella vectensis.[10] There are 270 nuclear receptors in the roundworm Caenorhabditis elegans alone,[11] 21 in the fruit fly and other insects,[12] 73 in zebrafish.[13] Humans, mice, and rats have respectively 48, 49, and 47 nuclear receptors each.[14]
Ligands

Ligands that bind to and activate nuclear receptors include lipophilic substances such as endogenous hormones, vitamins A and D, and xenobiotic hormones. Because the expression of a large number of genes is regulated by nuclear receptors, ligands that activate these receptors can have profound effects on the organism. Many of these regulated genes are associated with various diseases, which explains why the molecular targets of approximately 13% of U.S. Food and Drug Administration (FDA) approved drugs target nuclear receptors.[15]
A number of nuclear receptors, referred to as orphan receptors,[16] have no known (or at least generally agreed upon) endogenous ligands. Some of these receptors such as FXR, LXR, and PPAR bind a number of metabolic intermediates such as fatty acids, bile acids and/or sterols with relatively low affinity. These receptors hence may function as metabolic sensors. Other nuclear receptors, such as CAR and PXR appear to function as xenobiotic sensors up-regulating the expression of cytochrome P450 enzymes that metabolize these xenobiotics.[17]
Structure
Most nuclear receptors have molecular masses between 50,000 and 100,000 daltons.
Nuclear receptors are modular in structure and contain the following domains:[18][19]
- (A-B) N-terminal regulatory domain: Contains the activation function 1 (AF-1) whose action is independent of the presence of ligand.[20] The transcriptional activation of AF-1 is normally very weak, but it does synergize with AF-2 in the E-domain (see below) to produce a more robust upregulation of gene expression. The A-B domain is highly variable in sequence between various nuclear receptors.
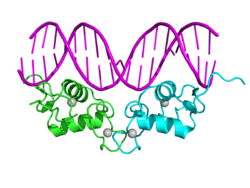
- (C) DNA-binding domain (DBD): Highly conserved domain containing two zinc fingers that binds to specific sequences of DNA called hormone response elements (HRE). Recently, a novel zinc finger motif (CHC2) is identified in parasitic flatworm NRs. [21]
- (D) Hinge region: Thought to be a flexible domain that connects the DBD with the LBD. Influences intracellular trafficking and subcellular distribution with a target peptide sequence.
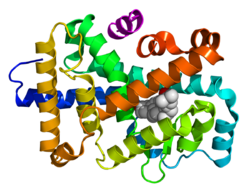
- (E) Ligand binding domain (LBD): Moderately conserved in sequence and highly conserved in structure between the various nuclear receptors. The structure of the LBD is referred to as an alpha helical sandwich fold in which three anti parallel alpha helices (the "sandwich filling") are flanked by two alpha helices on one side and three on the other (the "bread"). The ligand binding cavity is within the interior of the LBD and just below three anti parallel alpha helical sandwich "filling". Along with the DBD, the LBD contributes to the dimerization interface of the receptor and in addition, binds coactivator and corepressor proteins. The LBD also contains the activation function 2 (AF-2) whose action is dependent on the presence of bound ligand, controlled by the conformation of helix 12 (H12).[20]
- (F) C-terminal domain: Highly variable in sequence between various nuclear receptors.
The N-terminal (A/B), DNA-binding (C), and ligand binding (E) domains are independently well folded and structurally stable while the hinge region (D) and optional C-terminal (F) domains may be conformationally flexible and disordered.[22] Domains relative orientations are very different by comparing three known multi-domain crystal structures, two of them binding on DR1 (DBDs separated by 1 bp),[1][23] one binding on DR4 (by 4 bp).[24]
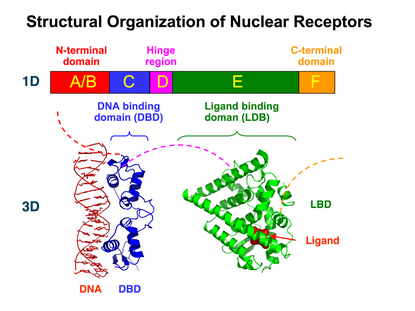 Top – Schematic 1D amino acid sequence of a nuclear receptor. Bottom – 3D structures of the DBD (bound to DNA) and LBD (bound to hormone) regions of the nuclear receptor. The structures shown are of the estrogen receptor. Experimental structures of N-terminal domain (A/B), hinge region (D), and C-terminal domain (F) have not been determined therefore are represented by red, purple, and orange dashed lines, respectively. |
|
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
Mechanism of action
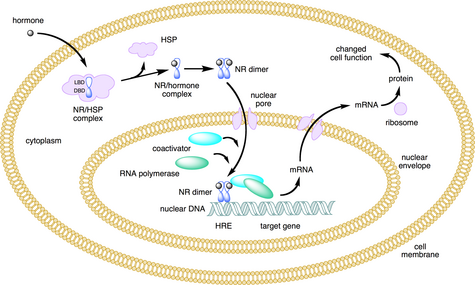
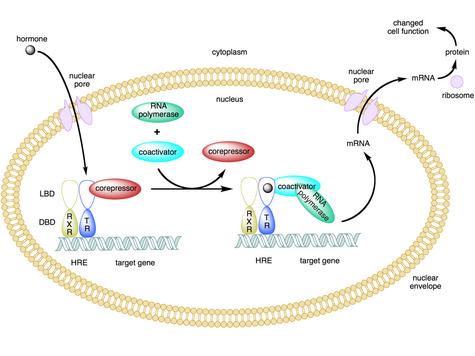
Nuclear receptors are multifunctional proteins that transduce signals of their cognate ligands. Nuclear receptors (NRs) may be classified into two broad classes according to their mechanism of action and subcellular distribution in the absence of ligand.
Small lipophilic substances such as natural hormones diffuse through the cell membrane and bind to nuclear receptors located in the cytosol (type I NR) or nucleus (type II NR) of the cell. Binding causes a conformational change in the receptor which, depending on the class of receptor, triggers a cascade of downstream events that direct the NRs to DNA transcription regulation sites which result in up or down-regulation of gene expression. They generally function as homo/heterodimers.[27] In addition, two additional classes, type III which are a variant of type I, and type IV that bind DNA as monomers have also been identified.[4]
Accordingly, nuclear receptors may be subdivided into the following four mechanistic classes:[4][5]
Type I
Ligand binding to type I nuclear receptors in the cytosol results in the dissociation of heat shock proteins, homo-dimerization, translocation (i.e., active transport) from the cytoplasm into the cell nucleus, and binding to specific sequences of DNA known as hormone response elements (HREs). Type I nuclear receptors bind to HREs consisting of two half-sites separated by a variable length of DNA, and the second half-site has a sequence inverted from the first (inverted repeat). Type I nuclear receptors include members of subfamily 3, such as the androgen receptor, estrogen receptors, glucocorticoid receptor, and progesterone receptor.[28]
It has been noted that some of the NR subfamily 2 nuclear receptors may bind to direct repeat instead of inverted repeat HREs. In addition, some nuclear receptors that bind either as monomers or dimers, with only a single DNA binding domain of the receptor attaching to a single half site HRE. These nuclear receptors are considered orphan receptors, as their endogenous ligands are still unknown.
The nuclear receptor/DNA complex then recruits other proteins that transcribe DNA downstream from the HRE into messenger RNA and eventually protein, which causes a change in cell function.
Type II
Type II receptors, in contrast to type I, are retained in the nucleus regardless of the ligand binding status and in addition bind as hetero-dimers (usually with RXR) to DNA.[27] In the absence of ligand, type II nuclear receptors are often complexed with corepressor proteins. Ligand binding to the nuclear receptor causes dissociation of corepressor and recruitment of coactivator proteins. Additional proteins including RNA polymerase are then recruited to the NR/DNA complex that transcribe DNA into messenger RNA.
Type II nuclear receptors include principally subfamily 1, for example the retinoic acid receptor, retinoid X receptor and thyroid hormone receptor.[29]
Type III
Type III nuclear receptors (principally NR subfamily 2) are similar to type I receptors in that both classes bind to DNA as homodimers. However, type III nuclear receptors, in contrast to type I, bind to direct repeat instead of inverted repeat HREs.
Type IV
Type IV nuclear receptors bind either as monomers or dimers, but only a single DNA binding domain of the receptor binds to a single half site HRE. Examples of type IV receptors are found in most of the NR subfamilies.
Dimerization
Human Nuclear Receptors are capable of dimerizing with many other Nuclear Receptors (homotypic dimerization), as has been shown from large-scale Y2H experiments and text mining efforts of the literature that were focused on specific interactions.[30][31][27] Nevertheless, there exists specificity, with members of the same subfamily having very similar NR dimerization partners and the underlying dimerization network has certain topological features, such as the presence of highly connected hubs (RXR and SHP).[27]
Coregulatory proteins
Nuclear receptors bound to hormone response elements recruit a significant number of other proteins (referred to as transcription coregulators) that facilitate or inhibit the transcription of the associated target gene into mRNA.[32][33][34] The function of these coregulators are varied and include chromatin remodeling (making the target gene either more or less accessible to transcription) or a bridging function to stabilize the binding of other coregulatory proteins. Nuclear receptors may bind specifically to a number of coregulator proteins, and thereby influence cellular mechanisms of signal transduction both directly, as well as indirectly.[35]
Coactivators
Binding of agonist ligands (see section below) to nuclear receptors induces a conformation of the receptor that preferentially binds coactivator proteins. These proteins often have an intrinsic histone acetyltransferase (HAT) activity, which weakens the association of histones to DNA, and therefore promotes gene transcription.
Corepressors
Binding of antagonist ligands to nuclear receptors in contrast induces a conformation of the receptor that preferentially binds corepressor proteins. These proteins, in turn, recruit histone deacetylases (HDACs), which strengthens the association of histones to DNA, and therefore represses gene transcription.
Agonism vs antagonism
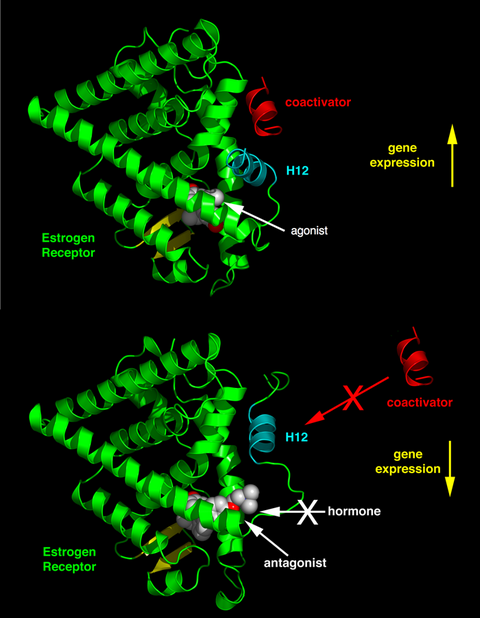
Depending on the receptor involved, the chemical structure of the ligand and the tissue that is being affected, nuclear receptor ligands may display dramatically diverse effects ranging in a spectrum from agonism to antagonism to inverse agonism.[38]
Agonists
The activity of endogenous ligands (such as the hormones estradiol and testosterone) when bound to their cognate nuclear receptors is normally to upregulate gene expression. This stimulation of gene expression by the ligand is referred to as an agonist response. The agonistic effects of endogenous hormones can also be mimicked by certain synthetic ligands, for example, the glucocorticoid receptor anti-inflammatory drug dexamethasone. Agonist ligands work by inducing a conformation of the receptor which favors coactivator binding (see upper half of the figure to the right).
Antagonists
Other synthetic nuclear receptor ligands have no apparent effect on gene transcription in the absence of endogenous ligand. However they block the effect of agonist through competitive binding to the same binding site in the nuclear receptor. These ligands are referred to as antagonists. An example of antagonistic nuclear receptor drug is mifepristone which binds to the glucocorticoid and progesterone receptors and therefore blocks the activity of the endogenous hormones cortisol and progesterone respectively. Antagonist ligands work by inducing a conformation of the receptor which prevents coactivator binding, and promotes corepressor binding (see lower half of the figure to the right).
Inverse agonists
Finally, some nuclear receptors promote a low level of gene transcription in the absence of agonists (also referred to as basal or constitutive activity). Synthetic ligands which reduce this basal level of activity in nuclear receptors are known as inverse agonists.[39]
Selective receptor modulators
A number of drugs that work through nuclear receptors display an agonist response in some tissues and an antagonistic response in other tissues. This behavior may have substantial benefits since it may allow retaining the desired beneficial therapeutic effects of a drug while minimizing undesirable side effects. Drugs with this mixed agonist/antagonist profile of action are referred to as selective receptor modulators (SRMs). Examples include Selective Androgen Receptor Modulators (SARMs), Selective Estrogen Receptor Modulators (SERMs) and Selective Progesterone Receptor Modulators (SPRMs). The mechanism of action of SRMs may vary depending on the chemical structure of the ligand and the receptor involved, however it is thought that many SRMs work by promoting a conformation of the receptor that is closely balanced between agonism and antagonism. In tissues where the concentration of coactivator proteins is higher than corepressors, the equilibrium is shifted in the agonist direction. Conversely in tissues where corepressors dominate, the ligand behaves as an antagonist.[40]
Alternative mechanisms
Transrepression
The most common mechanism of nuclear receptor action involves direct binding of the nuclear receptor to a DNA hormone response element. This mechanism is referred to as transactivation. However some nuclear receptors not only have the ability to directly bind to DNA, but also to other transcription factors. This binding often results in deactivation of the second transcription factor in a process known as transrepression.[41] One example of a nuclear receptor that are able to transrepress is the glucocorticoid receptor (GR). Furthermore, certain GR ligands known as Selective Glucocorticoid Receptor Agonists (SEGRAs) are able to activate GR in such a way that GR more strongly transrepresses than transactivates. This selectivity increases the separation between the desired antiinflammatory effects and undesired metabolic side effects of these selective glucocorticoids.
Non-genomic
The classical direct effects of nuclear receptors on gene regulation normally take hours before a functional effect is seen in cells because of the large number of intermediate steps between nuclear receptor activation and changes in protein expression levels. However it has been observed that many effects of the application of nuclear hormones, such as changes in ion channel activity, occur within minutes which is inconsistent with the classical mechanism of nuclear receptor action. While the molecular target for these non-genomic effects of nuclear receptors has not been conclusively demonstrated, it has been hypothesized that there are variants of nuclear receptors which are membrane associated instead of being localized in the cytosol or nucleus. Furthermore, these membrane associated receptors function through alternative signal transduction mechanisms not involving gene regulation.[42][43]
While it has been hypothesized that there are several membrane associated receptors for nuclear hormones, many of the rapid effects have been shown to require canonical nuclear receptors.[44][45] However, testing the relative importance of the genomic and nongenomic mechanisms in vivo has been prevented by the absence of specific molecular mechanisms for the nongenomic effects that could be blocked by mutation of the receptor without disrupting its direct effects on gene expression.
A molecular mechanism for non-genomic signaling through the nuclear thyroid hormone receptor TRβ involves the phosphatidylinositol 3-kinase (PI3K).[46] This signaling can be blocked by a single tyrosine to phenylalanine substitution in TRβ without disrupting direct gene regulation.[47] When mice were created with this single, conservative amino acid substitution in TRβ,[47] synaptic maturation and plasticity in the hippocampus was impaired almost as effectively as completely blocking thyroid hormone synthesis.[48] This mechanism appears to be conserved in all mammals but not in TRα or any other nuclear receptors. Thus, phosphotyrosine-dependent association of TRβ with PI3K provides a potential mechanism for integrating regulation of development and metabolism by thyroid hormone and receptor tyrosine kinases. In addition, thyroid hormone signaling through PI3K can alter gene expression.[49]
Family members
The following is a list of the 48 known human nuclear receptors (and their orthologs in other species)[14][50][51] categorized according to sequence homology.[6][7] The list also includes selected family members that lack human orthologs (NRNC symbol highlighted in yellow).
| Subfamily | Group | Member | ||||||
|---|---|---|---|---|---|---|---|---|
| NRNC Symbol[6] | Abbreviation | Name | Gene | Ligand(s) | ||||
| 1 | Thyroid Hormone Receptor-like | A | Thyroid hormone receptor | NR1A1 | TRα | Thyroid hormone receptor-α | THRA | thyroid hormone |
| NR1A2 | TRβ | Thyroid hormone receptor-β | THRB | |||||
| B | Retinoic acid receptor | NR1B1 | RARα | Retinoic acid receptor-α | RARA | vitamin A and related compounds | ||
| NR1B2 | RARβ | Retinoic acid receptor-β | RARB | |||||
| NR1B3 | RARγ | Retinoic acid receptor-γ | RARG | |||||
| C | Peroxisome proliferator-activated receptor | NR1C1 | PPARα | Peroxisome proliferator-activated receptor-α | PPARA | fatty acids, prostaglandins | ||
| NR1C2 | PPAR-β/δ | Peroxisome proliferator-activated receptor-β/δ | PPARD | |||||
| NR1C3 | PPARγ | Peroxisome proliferator-activated receptor-γ | PPARG | |||||
| D | Rev-ErbA | NR1D1 | Rev-ErbAα | Rev-ErbAα | NR1D1 | heme | ||
| NR1D2 | Rev-ErbAβ | Rev-ErbAα | NR1D2 | |||||
| E | E78C-like (arthropod, trematode, mullosc, nematode)[50][52] |
NR1E1 | Eip78C | Ecdysone-induced protein 78C | Eip78C | |||
| F | RAR-related orphan receptor | NR1F1 | RORα | RAR-related orphan receptor-α | RORA | cholesterol, ATRA | ||
| NR1F2 | RORβ | RAR-related orphan receptor-β | RORB | |||||
| NR1F3 | RORγ | RAR-related orphan receptor-γ | RORC | |||||
| G | CNR14-like (nematode)[50] | NR1G1 | sex-1 | Steroid hormone receptor cnr14[53] | sex-1 | |||
| H | Liver X receptor-like | NR1H1 | EcR | Ecdysone receptor, EcR (arthropod) | EcR | ecdysteroids | ||
| NR1H2 | LXRβ | Liver X receptor-β | NR1H2 | oxysterols | ||||
| NR1H3 | LXRα | Liver X receptor-α | NR1H3 | |||||
| NR1H4 | FXR | Farnesoid X receptor | NR1H4 | |||||
| NR1H5[54] | FXR-β | Farnesoid X receptor-β (pseudogene in human) |
NR1H5P | |||||
| I | Vitamin D receptor-like | NR1I1 | VDR | Vitamin D receptor | VDR | vitamin D | ||
| NR1I2 | PXR | Pregnane X receptor | NR1I2 | xenobiotics | ||||
| NR1I3 | CAR | Constitutive androstane receptor | NR1I3 | androstane | ||||
| J | Hr96-like[50] | NR1J1 | Hr96/Daf-12 | Nuclear hormone receptor HR96 | Hr96 | cholesterol/dafachronic acid[55] | ||
| NR1J2 | ||||||||
| NR1J3 | ||||||||
| K | Hr1-like[50] | NR1K1 | Hr1 | Nuclear hormone receptor HR1 | ||||
| 2 | Retinoid X Receptor-like | A | Hepatocyte nuclear factor-4 | NR2A1 | HNF4α | Hepatocyte nuclear factor-4-α | HNF4A | fatty acids |
| NR2A2 | HNF4γ | Hepatocyte nuclear factor-4-γ | HNF4G | |||||
| B | Retinoid X receptor | NR2B1 | RXRα | Retinoid X receptor-α | RXRA | retinoids | ||
| NR2B2 | RXRβ | Retinoid X receptor-β | RXRB | |||||
| NR2B3 | RXRγ | Retinoid X receptor-γ | RXRG | |||||
| NR2B4 | USP | Ultraspiracle protein (arthropod) | usp | phospholipids[56] | ||||
| C | Testicular receptor | NR2C1 | TR2 | Testicular receptor 2 | NR2C1 | |||
| NR2C2 | TR4 | Testicular receptor 4 | NR2C2 | |||||
| E | TLX/PNR | NR2E1 | TLX | Homologue of the Drosophila tailless gene | NR2E1 | |||
| NR2E3 | PNR | Photoreceptor cell-specific nuclear receptor | NR2E3 | |||||
| F | COUP/EAR | NR2F1 | COUP-TFI | Chicken ovalbumin upstream promoter-transcription factor I | NR2F1 | |||
| NR2F2 | COUP-TFII | Chicken ovalbumin upstream promoter-transcription factor II | NR2F2 | retinoic acid (weak)[57] | ||||
| NR2F6 | EAR-2 | V-erbA-related | NR2F6 | |||||
| 3 | Estrogen Receptor-like | A | Estrogen receptor | NR3A1 | ERα | Estrogen receptor-α | ESR1 | estrogens |
| NR3A2 | ERβ | Estrogen receptor-β | ESR2 | |||||
| B | Estrogen related receptor | NR3B1 | ERRα | Estrogen-related receptor-α | ESRRA | |||
| NR3B2 | ERRβ | Estrogen-related receptor-β | ESRRB | |||||
| NR3B3 | ERRγ | Estrogen-related receptor-γ | ESRRG | |||||
| C | 3-Ketosteroid receptors | NR3C1 | GR | Glucocorticoid receptor | NR3C1 | cortisol | ||
| NR3C2 | MR | Mineralocorticoid receptor | NR3C2 | aldosterone | ||||
| NR3C3 | PR | Progesterone receptor | PGR | progesterone | ||||
| NR3C4 | AR | Androgen receptor | AR | testosterone | ||||
| D | Estrogen Receptor-like (in lophotrochozoa)[58] |
NR3D | ||||||
| E | Estrogen Receptor-like (in cnidaria)[59] |
NR3E | ||||||
| F | Estrogen Receptor-like (in placozoa)[59] |
NR3F | ||||||
| 4 | Nerve Growth Factor IB-like | A | NGFIB/NURR1/NOR1 | NR4A1 | NGFIB | Nerve Growth factor IB | NR4A1 | |
| NR4A2 | NURR1 | Nuclear receptor related 1 | NR4A2 | |||||
| NR4A3 | NOR1 | Neuron-derived orphan receptor 1 | NR4A3 | |||||
| 5 | Steroidogenic Factor-like |
A | SF1/LRH1 | NR5A1 | SF1 | Steroidogenic factor 1 | NR5A1 | phosphatidylinositols |
| NR5A2 | LRH-1 | Liver receptor homolog-1 | NR5A2 | phosphatidylinositols | ||||
| B | Hr39-like | NR5B1[50] | HR39/FTZ-F1 | Nuclear hormone receptor fushi tarazu factor I beta | Hr39 | |||
| 6 | Germ Cell Nuclear Factor-like | A | GCNF | NR6A1 | GCNF | Germ cell nuclear factor | NR6A1 | |
| 7 | NRs with two DNA binding domains[60][50][61] | A | 2DBD-NRα | NR7A1 | 2DBD-NRA2 | |||
| B | 2DBD-NRβ | NR7B1 | 2DBD-NRA3 | |||||
| C | 2DBD-NRγ | NR7C1 | 2DBD-NRA1 | arthropod "α/β" | ||||
| 8 | NR8[62] (eumetazoa) | A | NR8A | NR8A1 | CgNR8A1 | Nuclear receptor 8 | AKG49571 | |
| 0 | Miscellaneous (lacks either LBD or DBD) | A | knr/knrl/egon[50] (arthropods) | NR0A1 | KNI | Zygotic gap protein knirps | knl | |
| B | DAX/SHP | NR0B1 | DAX1 | Dosage-sensitive sex reversal, adrenal hypoplasia critical region, on chromosome X, gene 1 | NR0B1 | |||
| NR0B2 | SHP | Small heterodimer partner | NR0B2 | |||||
Of the two 0-families, 0A has a family 1-like DBD, and 0B has a very unique LBD. The second DBD of family 7 is probably related to the family 1 DBD. Three probably family-1 NRs from Biomphalaria glabrata possess a DBD along with an family 0B-like LBD.[50] The placement of C. elegans nhr-1 (Q21878) is disputed: although most sources place it as NR1K1,[50] manual annotation at WormBase considers it a member of NR2A.[63] There used to be a group 2D for which the only member was Drosophila HR78/NR1D1 (Q24142) and orthologues, but it was merged into group 2C later due to high similarity, forming a "group 2C/D".[50] Knockout studies on mice and fruit flies support such a merged group.[64]
Evolution
A topic of debate has been on the identity of the ancestral nuclear receptor as either a ligand-binding or an orphan receptor. This debate began more than twenty-five years ago when the first ligands were identified as mammalian steroid and thyroid hormones.[65] Shortly thereafter, the identification of the ecdysone receptor in Drosophila introduced the idea that nuclear receptors were hormonal receptors that bind ligands with a nanomolar affinity. At the time, the three known nuclear receptor ligands were steroids, retinoids, and thyroid hormone, and of those three, both steroids and retinoids were products of terpenoid metabolism. Thus, it was postulated that ancestral receptor would have been liganded by a terpenoid molecule.[66]
In 1992, a comparison of the DNA-binding domain of all known nuclear receptors led to the construction of a phylogenic tree of nuclear receptor that indicated that all nuclear receptors shared a common ancestor.[67] As a result, there was an increased effort upon uncovering the state of the first nuclear receptor, and by 1997 an alternative hypothesis was suggested: the ancestral nuclear receptor was an orphan receptor and it acquired ligand-binding ability over time[7] This hypothesis was proposed based on the following arguments:
- The nuclear receptor sequences that had been identified in the earliest metazoans (cnidarians and Schistosoma) were all members of the COUP-TF, RXR, and FTZ-F1 groups of receptors. Both COUP-TF and FTZ-F1 are orphan receptors, and RXR is only found to bind a ligand in vertebrates.[68]
- While orphan receptors had known arthropod homologs, no orthologs of liganded vertebrate receptors had been identified outside vertebrates, suggesting that orphan receptors are older than liganded-receptors.[69]
- Orphan receptors are found amongst all six subfamilies of nuclear receptors, while ligand-dependent receptors are found amongst three.[7] Thus, since the ligand-dependent receptors were believed to be predominantly member of recent subfamilies, it seemed logical that they gained the ability to bind ligands independently.
- The phylogenetic position of a given nuclear receptor within the tree correlates to its DNA-binding domain and dimerization abilities, but there is no identified relationship between a ligand-dependent nuclear receptor and the chemical nature of its ligand. In addition to this, the evolutionary relationships between ligand-dependent receptors did not make much sense as closely related receptors of subfamilies bound ligands originating from entirely different biosynthetic pathways (e.g. TRs and RARs). On the other hand, subfamilies that are not evolutionarily related bind similar ligands (RAR and RXR both bind all-trans and 9-cis retinoic acid respectively).[69]
- In 1997, it was discovered that nuclear receptors did not exist in static off and on conformations, but that a ligand could alter the equilibrium between the two states. Furthermore, it was found that nuclear receptors could be regulated in a ligand-independent manner, through either phosphorylation or other post-translational modifications. Thus, this provided a mechanism for how an ancestral orphan receptor was regulated in a ligand-independent manner, and explained why the ligand binding domain was conserved.[69]
Over the next 10 years, experiments were conducted to test this hypothesis and counterarguments soon emerged:
- Nuclear receptors were identified in the newly sequenced genome of the demosponge Amphimedon queenslandica, a member Porifera, the most ancient metazoan phylum. The A. queenslandica genome contains two nuclear receptors known as AqNR1 and AqNR2 and both were characterized to bind and be regulated by ligands.[70]
- Homologs for ligand-dependent vertebrate receptors were found outside vertebrates in mollusks and Platyhelminthes. Furthermore, the nuclear receptors found in cnidarians were found to have structural ligands in mammals, which could mirror the ancestral situation.
- Two putative orphan receptors, HNF4 and USP were found, via structural and mass spectrometry analysis, to bind fatty acids and phospholipids respectively.[56]
- Nuclear receptors and ligands are found to be a lot less specific than was previously thought. Retinoids can bind mammalian receptors other than RAR and RXR such as, PPAR, RORb, or COUP-TFII. Furthermore, RXR is sensitive to a wide range of molecules including retinoids, fatty acids, and phospholipids.[71]
- Study of steroid receptor evolution revealed that the ancestral steroid receptor could bind a ligand, estradiol. Conversely, the estrogen receptor found in mollusks is constitutively active and did not bind estrogen-related hormones. Thus, this provided an example of how an ancestral ligand-dependent receptor could lose its ability to bind ligands.[72]
A combination of this recent evidence, as well as an in-depth study of the physical structure of the nuclear receptor ligand binding domain has led to the emergence of a new hypothesis regarding the ancestral state of the nuclear receptor. This hypothesis suggests that the ancestral receptor may act as a lipid sensor with an ability to bind, albeit rather weakly, several different hydrophobic molecules such as, retinoids, steroids, hemes, and fatty acids. With its ability to interact with a variety of compounds, this receptor, through duplications, would either lose its ability for ligand-dependent activity, or specialize into a highly specific receptor for a particular molecule.[71]
History
Below is a brief selection of key events in the history of nuclear receptor research.[73]
- 1905 – Ernest Starling coined the word hormone
- 1926 – Edward Calvin Kendall and Tadeus Reichstein isolated and determined the structures of cortisone and thyroxine
- 1929 – Adolf Butenandt and Edward Adelbert Doisy – independently isolated and determined the structure of estrogen
- 1958 – Elwood Jensen – isolated the estrogen receptor
- 1980s – cloning of the estrogen, glucocorticoid, and thyroid hormone receptors by Pierre Chambon, Ronald Evans, and Björn Vennström respectively
- 2004 – Pierre Chambon, Ronald Evans, and Elwood Jensen were awarded the Albert Lasker Award for Basic Medical Research, an award that frequently precedes a Nobel Prize in Medicine
See also
- NucleaRDB
- Thyroid hormone receptor
- Steroid hormone receptor
- Receptors
References
- ↑ 1.0 1.1 PDB: 3E00; "Structure of the intact PPAR-gamma-RXR- nuclear receptor complex on DNA". Nature 456 (7220): 350–6. November 2008. doi:10.1038/nature07413. PMID 19043829.
- ↑ "The steroid and thyroid hormone receptor superfamily". Science 240 (4854): 889–95. May 1988. doi:10.1126/science.3283939. PMID 3283939. Bibcode: 1988Sci...240..889E.
- ↑ "Nuclear receptor minireview series". The Journal of Biological Chemistry 276 (40): 36863–4. October 2001. doi:10.1074/jbc.R100047200. PMID 11459855.
- ↑ 4.0 4.1 4.2 "The nuclear receptor superfamily: the second decade". Cell 83 (6): 835–9. December 1995. doi:10.1016/0092-8674(95)90199-X. PMID 8521507.
- ↑ 5.0 5.1 "Nuclear receptors: overview and classification". Current Drug Targets. Inflammation and Allergy 3 (4): 335–46. December 2004. doi:10.2174/1568010042634541. PMID 15584884.
- ↑ 6.0 6.1 6.2 Nuclear Receptors Nomenclature Committee (April 1999). "A unified nomenclature system for the nuclear receptor superfamily". Cell 97 (2): 161–3. doi:10.1016/S0092-8674(00)80726-6. PMID 10219237.
- ↑ 7.0 7.1 7.2 7.3 "Evolution of the nuclear receptor superfamily: early diversification from an ancestral orphan receptor". Journal of Molecular Endocrinology 19 (3): 207–26. December 1997. doi:10.1677/jme.0.0190207. PMID 9460643.
- ↑ "Evolution and diversification of the nuclear receptor superfamily". Annals of the New York Academy of Sciences 839 (1): 143–6. May 1998. doi:10.1111/j.1749-6632.1998.tb10747.x. PMID 9629140. Bibcode: 1998NYASA.839..143E.
- ↑ "Nuclear receptors from the ctenophore Mnemiopsis leidyi lack a zinc-finger DNA-binding domain: lineage-specific loss or ancestral condition in the emergence of the nuclear receptor superfamily?". EvoDevo 2 (1): 3. February 2011. doi:10.1186/2041-9139-2-3. PMID 21291545.
- ↑ "Protein evolution by molecular tinkering: diversification of the nuclear receptor superfamily from a ligand-dependent ancestor". PLOS Biology 8 (10): e1000497. October 2010. doi:10.1371/journal.pbio.1000497. PMID 20957188.
- ↑ "Nuclear receptors in nematodes: themes and variations". Trends in Genetics 17 (4): 206–13. April 2001. doi:10.1016/S0168-9525(01)02242-9. PMID 11275326.
- ↑ "The Function and Evolution of Nuclear Receptors in Insect Embryonic Development". Current Topics in Developmental Biology 125: 39–70. 2017. doi:10.1016/bs.ctdb.2017.01.003. ISBN 9780128021729. PMID 28527580.
- ↑ "Nuclear receptor research in zebrafish". Journal of Molecular Endocrinology 59 (1): R65–R76. 2017. doi:10.1530/JME-17-0031. PMID 28438785.
- ↑ 14.0 14.1 "Genomic analysis of the nuclear receptor family: new insights into structure, regulation, and evolution from the rat genome". Genome Research 14 (4): 580–90. April 2004. doi:10.1101/gr.2160004. PMID 15059999.
- ↑ "How many drug targets are there?". Nature Reviews. Drug Discovery 5 (12): 993–6. December 2006. doi:10.1038/nrd2199. PMID 17139284.
- ↑ "International Union of Pharmacology. LXVI. Orphan nuclear receptors". Pharmacological Reviews 58 (4): 798–836. December 2006. doi:10.1124/pr.58.4.10. PMID 17132856.
- ↑ "Orphan nuclear receptor modulators". Current Topics in Medicinal Chemistry 3 (14): 1637–47. 2003. doi:10.2174/1568026033451709. PMID 14683519.
- ↑ "The structure of the nuclear hormone receptors". Steroids 64 (5): 310–9. May 1999. doi:10.1016/S0039-128X(99)00014-8. PMID 10406480.
- ↑ "Estrogen receptor interaction with co-activators and co-repressors". Steroids 65 (5): 227–51. May 2000. doi:10.1016/S0039-128X(99)00107-5. PMID 10751636.
- ↑ 20.0 20.1 "Activation functions 1 and 2 of nuclear receptors: molecular strategies for transcriptional activation". Molecular Endocrinology 17 (10): 1901–9. October 2003. doi:10.1210/me.2002-0384. PMID 12893880.
- ↑ "Identification and evolution of nuclear receptors in Platyhelminths". PLOS ONE 16(8): e0250750 (8): e0250750. 2021. doi:10.1371/journal.pone.0250750. PMID 34388160. Bibcode: 2021PLoSO..1650750W.
- ↑ "Nuclear-receptor ligands and ligand-binding domains". Annual Review of Biochemistry 68: 559–81. 1999. doi:10.1146/annurev.biochem.68.1.559. PMID 10872460.
- ↑ "Multidomain integration in the structure of the HNF-4α nuclear receptor complex". Nature 495 (7441): 394–8. March 2013. doi:10.1038/nature11966. PMID 23485969. Bibcode: 2013Natur.495..394C.
- ↑ "Structure of the retinoid X receptor α-liver X receptor β (RXRα-LXRβ) heterodimer on DNA". Nature Structural & Molecular Biology 21 (3): 277–81. March 2014. doi:10.1038/nsmb.2778. PMID 24561505.
- ↑ PDB: 2C7A; "Structure of the progesterone receptor-deoxyribonucleic acid complex: novel interactions required for binding to half-site response elements". Molecular Endocrinology 20 (12): 3042–52. December 2006. doi:10.1210/me.2005-0511. PMID 16931575.
- ↑ PDB: 3L0L; "Structural basis for hydroxycholesterols as natural ligands of orphan nuclear receptor RORgamma". Molecular Endocrinology 24 (5): 923–9. May 2010. doi:10.1210/me.2009-0507. PMID 20203100.
- ↑ 27.0 27.1 27.2 27.3 "A protein interaction atlas for the nuclear receptors: properties and quality of a hub-based dimerisation network". BMC Systems Biology 1: 34. July 2007. doi:10.1186/1752-0509-1-34. PMID 17672894.
- ↑ "Expression of androgen receptor coregulators in prostate cancer". Clinical Cancer Research 10 (3): 1032–40. February 2004. doi:10.1158/1078-0432.CCR-0990-3. PMID 14871982.
- ↑ "Binding of type II nuclear receptors and estrogen receptor to full and half-site estrogen response elements in vitro". Nucleic Acids Research 25 (10): 1903–12. May 1997. doi:10.1093/nar/25.10.1903. PMID 9115356.
- ↑ Rual, Jean-François; Venkatesan, Kavitha; Hao, Tong; Hirozane-Kishikawa, Tomoko; Dricot, Amélie; Li, Ning; Berriz, Gabriel F.; Gibbons, Francis D. et al. (2005-10-20). "Towards a proteome-scale map of the human protein-protein interaction network". Nature 437 (7062): 1173–1178. doi:10.1038/nature04209. ISSN 1476-4687. PMID 16189514. Bibcode: 2005Natur.437.1173R. https://pubmed.ncbi.nlm.nih.gov/16189514.
- ↑ Albers, Michael; Kranz, Harald; Kober, Ingo; Kaiser, Carmen; Klink, Martin; Suckow, Jörg; Kern, Rainer; Koegl, Manfred (February 2005). "Automated yeast two-hybrid screening for nuclear receptor-interacting proteins". Molecular & Cellular Proteomics 4 (2): 205–213. doi:10.1074/mcp.M400169-MCP200. ISSN 1535-9476. PMID 15604093.
- ↑ "Nuclear receptor coregulators: cellular and molecular biology". Endocrine Reviews 20 (3): 321–344. June 1999. doi:10.1210/edrv.20.3.0366. PMID 10368774.
- ↑ "The coregulator exchange in transcriptional functions of nuclear receptors". Genes & Development 14 (2): 121–41. January 2000. doi:10.1101/gad.14.2.121. PMID 10652267. http://genesdev.cshlp.org/content/14/2/121.long.
- ↑ "Nuclear hormone receptors and gene expression". Physiological Reviews 81 (3): 1269–304. July 2001. doi:10.1152/physrev.2001.81.3.1269. PMID 11427696.
- ↑ "Sex steroid receptors in skeletal differentiation and epithelial neoplasia: is tissue-specific intervention possible?". BioEssays 31 (6): 629–41. June 2009. doi:10.1002/bies.200800138. PMID 19382224.
- ↑ "Molecular basis of agonism and antagonism in the oestrogen receptor". Nature 389 (6652): 753–8. October 1997. doi:10.1038/39645. PMID 9338790. Bibcode: 1997Natur.389..753B.
- ↑ "The structural basis of estrogen receptor/coactivator recognition and the antagonism of this interaction by tamoxifen". Cell 95 (7): 927–37. December 1998. doi:10.1016/S0092-8674(00)81717-1. PMID 9875847.
- ↑ "Principles for modulation of the nuclear receptor superfamily". Nature Reviews. Drug Discovery 3 (11): 950–64. November 2004. doi:10.1038/nrd1551. PMID 15520817.
- ↑ "Identification of a selective inverse agonist for the orphan nuclear receptor estrogen-related receptor alpha". Journal of Medicinal Chemistry 47 (23): 5593–6. November 2004. doi:10.1021/jm049334f. PMID 15509154.
- ↑ "Coregulator function: a key to understanding tissue specificity of selective receptor modulators". Endocrine Reviews 25 (1): 45–71. February 2004. doi:10.1210/er.2003-0023. PMID 14769827.
- ↑ "Nuclear receptors versus inflammation: mechanisms of transrepression". Trends in Endocrinology and Metabolism 17 (8): 321–7. October 2006. doi:10.1016/j.tem.2006.08.005. PMID 16942889.
- ↑ "Estrogen receptor-dependent activation of AP-1 via non-genomic signalling". Nuclear Receptor 2 (1): 3. June 2004. doi:10.1186/1478-1336-2-3. PMID 15196329.
- ↑ "Membrane estrogen receptor-alpha levels in MCF-7 breast cancer cells predict cAMP and proliferation responses". Breast Cancer Research 7 (1): R101–12. 2005. doi:10.1186/bcr958. PMID 15642158.
- ↑ "Nongenotropic, sex-nonspecific signaling through the estrogen or androgen receptors: dissociation from transcriptional activity". Cell 104 (5): 719–30. March 2001. doi:10.1016/S0092-8674(01)00268-9. PMID 11257226.
- ↑ "Rapid signaling at the plasma membrane by a nuclear receptor for thyroid hormone". Proceedings of the National Academy of Sciences of the United States of America 103 (13): 5197–201. March 2006. doi:10.1073/pnas.0600089103. PMID 16549781. Bibcode: 2006PNAS..103.5197S.
- ↑ "Rac and Rho mediate opposing hormonal regulation of the ether-a-go-go-related potassium channel". Current Biology 12 (1): 27–33. January 2002. doi:10.1016/S0960-9822(01)00625-X. PMID 11790300.
- ↑ 47.0 47.1 "A rapid cytoplasmic mechanism for PI3 kinase regulation by the nuclear thyroid hormone receptor, TRβ, and genetic evidence for its role in the maturation of mouse hippocampal synapses in vivo". Endocrinology 155 (9): 3713–24. September 2014. doi:10.1210/en.2013-2058. PMID 24932806.
- ↑ "Alterations in synaptic transmission and plasticity in area CA1 of adult hippocampus following developmental hypothyroidism". Brain Research. Developmental Brain Research 148 (1): 11–8. January 2004. doi:10.1016/j.devbrainres.2003.09.018. PMID 14757514. https://zenodo.org/record/1258893.
- ↑ "Transcriptional regulation by nonclassical action of thyroid hormone". Thyroid Research 4 (Suppl 1): S6. August 2011. doi:10.1186/1756-6614-4-S1-S6. PMID 21835053.
- ↑ 50.00 50.01 50.02 50.03 50.04 50.05 50.06 50.07 50.08 50.09 50.10 "The nuclear receptors of Biomphalaria glabrata and Lottia gigantea: implications for developing new model organisms". PLOS ONE 10 (4): e0121259. 7 April 2015. doi:10.1371/journal.pone.0121259. PMID 25849443. Bibcode: 2015PLoSO..1021259K.
- ↑ Ohlstein, Eliot, ed (November 2023). "International Union of Basic and Clinical Pharmacology CXIII: Nuclear Receptor Superfamily—Update 2023" (in en). Pharmacological Reviews 75 (6): 1233–1318. doi:10.1124/pharmrev.121.000436. ISSN 0031-6997. PMID 37586884.
- ↑ "Dirofilaria immitis encodes Di-nhr-7, a putative orthologue of the Drosophila ecdysone-regulated E78 gene". Molecular and Biochemical Parasitology 119 (2): 169–77. February 2002. doi:10.1016/s0166-6851(01)00412-1. PMID 11814569.
- ↑ "sex-1 (gene)". https://www.wormbase.org/species/c_elegans/gene/WBGene00004786.
- ↑ "Identification of farnesoid X receptor beta as a novel mammalian nuclear receptor sensing lanosterol". Molecular and Cellular Biology 23 (3): 864–72. February 2003. doi:10.1128/mcb.23.3.864-872.2003. PMID 12529392.
- ↑ "FlyBase Gene Report: Dmel\Hr96". http://flybase.org/reports/FBgn0015240.html.
- ↑ 56.0 56.1 "Nuclear receptors: the evolution of diversity". Science's STKE 2004 (217): pe4. January 2004. doi:10.1126/stke.2172004pe4. PMID 14747695.
- ↑ "Identification of COUP-TFII orphan nuclear receptor as a retinoic acid-activated receptor". PLOS Biology 6 (9): e227. September 2008. doi:10.1371/journal.pbio.0060227. PMID 18798693.
- ↑ "Origin of an ancient hormone/receptor couple revealed by resurrection of an ancestral estrogen". Science Advances 3 (3): e1601778. March 2017. doi:10.1126/sciadv.1601778. PMID 28435861. Bibcode: 2017SciA....3E1778M.
- ↑ 59.0 59.1 "NR3E receptors in cnidarians : a new family of steroid receptor relatives extends the possible mechanisms for ligand binding". J Steroid Biochem Mol Biol 184: 11–19. November 2018. doi:10.1016/j.jsbmb.2018.06.014. PMID 29940311.
- ↑ "Updated knowledge and a proposed nomenclature for nuclear receptors with two DNA binding domains (2DBD-NRs)". PLOS ONE 18 (9): e0286107. September 2023. doi:10.1371/journal.pone.0286107. PMID 37699039. Bibcode: 2023PLoSO..1886107W.
- ↑ "Evolution of a novel subfamily of nuclear receptors with members that each contain two DNA binding domains". BMC Evol Biol 7 (27): 27. February 2007. doi:10.1186/1471-2148-7-27. PMID 17319953.
- ↑ "Evolution of a novel nuclear receptor subfamily with emphasis on the member from the Pacific oyster Crassostrea gigas". Gene 567 (2): 164–72. August 2015. doi:10.1016/j.gene.2015.04.082. PMID 25956376.
- ↑ "nhr-1 (gene)". https://www.wormbase.org/species/c_elegans/gene/WBGene00003600.
- ↑ "Adult functions for the Drosophila DHR78 nuclear receptor". Developmental Dynamics 247 (2): 315–322. February 2018. doi:10.1002/dvdy.24608. PMID 29171103.
- ↑ "The steroid and thyroid hormone receptor superfamily". Science 240 (4854): 889–95. May 1988. doi:10.1126/science.3283939. PMID 3283939. Bibcode: 1988Sci...240..889E.
- ↑ "Diversity and unity in the nuclear hormone receptors: a terpenoid receptor superfamily". The New Biologist 2 (1): 100–5. January 1990. PMID 1964083.
- ↑ "Evolution of the nuclear receptor gene superfamily". The EMBO Journal 11 (3): 1003–13. March 1992. doi:10.1002/j.1460-2075.1992.tb05139.x. PMID 1312460.
- ↑ "Ligand binding was acquired during evolution of nuclear receptors". Proceedings of the National Academy of Sciences of the United States of America 94 (13): 6803–8. June 1997. doi:10.1073/pnas.94.13.6803. PMID 9192646. Bibcode: 1997PNAS...94.6803E.
- ↑ 69.0 69.1 69.2 "Ligand binding and nuclear receptor evolution". BioEssays 22 (8): 717–27. August 2000. doi:10.1002/1521-1878(200008)22:8<717::AID-BIES5>3.0.CO;2-I. PMID 10918302.
- ↑ "Protein evolution by molecular tinkering: diversification of the nuclear receptor superfamily from a ligand-dependent ancestor". PLOS Biology 8 (10): e1000497. October 2010. doi:10.1371/journal.pbio.1000497. PMID 20957188.
- ↑ 71.0 71.1 "Origin and evolution of the ligand-binding ability of nuclear receptors". Molecular and Cellular Endocrinology. Evolution of Nuclear Hormone Receptors 334 (1–2): 21–30. March 2011. doi:10.1016/j.mce.2010.10.017. PMID 21055443.
- ↑ "Resurrecting the ancestral steroid receptor: ancient origin of estrogen signaling". Science 301 (5640): 1714–7. September 2003. doi:10.1126/science.1086185. PMID 14500980. Bibcode: 2003Sci...301.1714T.
- ↑ "One hundred years of hormones". EMBO Reports 6 (6): 490–6. June 2005. doi:10.1038/sj.embor.7400444. PMID 15940278.
External links
- Nuclear+Receptors at the US National Library of Medicine Medical Subject Headings (MeSH)
- Vincent Laudet (2006). "The IUPHAR Compendium of the Pharmacology and Classification of the Nuclear Receptor Superfamily 2006E". Nuclear Receptor Compendium. The International Union of Basic and Clinical Pharmacology. http://www.iuphar-db.org/index_nh.jsp.
- "Nuclear Receptor online journal". Home page. published by BioMed Central (no longer accepting submissions since May 2007). http://www.nuclear-receptor.com.
- "Nuclear Receptor Resource". Georgetown University. http://nrr.georgetown.edu/NRR/nrrhome.htm.
- "Nuclear Receptor Signaling Atlas (Receptors, Coactivators, Corepressors and Ligands)". The NURSA Consortium. http://www.nursa.org/. "an NIH-funded research consortium and database; includes open-access PubMed-indexed journal, Nuclear Receptor Signaling"
- "Nuclear Receptor Resource". Jack Vanden Heuvel. http://www.nrresource.org/.
Template:Receptor/signaling modulators
 |