Biology:Aryl hydrocarbon receptor
 Generic protein structure example |
The aryl hydrocarbon receptor (also known as AhR, AHR, ahr, ahR, AH receptor, or dioxin receptor) is a protein that in humans is encoded by the AHR gene. The aryl hydrocarbon receptor is a transcription factor that regulates gene expression. It was originally thought to function primarily as a sensor of xenobiotic chemicals and also as the regulator of enzymes such as cytochrome P450s that metabolize these chemicals. The most notable of these xenobiotic chemicals are aromatic (aryl) hydrocarbons from which the receptor derives its name.
More recently, it has been discovered that AhR is activated (or deactivated) by a number of endogenous indole derivatives such as kynurenine. In addition to regulating metabolism enzymes, the AhR has roles in regulating immunity, stem cell maintenance, and cellular differentiation.[1][2][3]
The aryl hydrocarbon receptor is a member of the family of basic helix-loop-helix transcription factors. AhR binds several exogenous ligands such as natural plant flavonoids, polyphenols and indoles, as well as synthetic polycyclic aromatic hydrocarbons and dioxin-like compounds. AhR is a cytosolic transcription factor that is normally inactive, bound to several co-chaperones. Upon ligand binding to chemicals such as 2,3,7,8-tetrachlorodibenzo-p-dioxin (TCDD), the chaperones dissociate resulting in AhR translocating into the nucleus and dimerizing with ARNT (AhR nuclear translocator), leading to changes in gene transcription.
Protein functional domains

The AhR protein contains several domains critical for function and is classified as a member of the basic helix-loop-helix/Per-Arnt-Sim (bHLH/PAS) family of transcription factors.[4][5] The bHLH motif is located in the N-terminal of the protein and is a common entity in a variety of transcription factors.[6] Members of the bHLH superfamily have two functionally distinctive and highly conserved domains. The first is the basic-region (b), which is involved in the binding of the transcription factor to DNA. The second is the helix-loop-helix (HLH) region, which facilitates protein-protein interactions. Also contained with the AhR are two PAS domains, PAS-A and PAS-B, which are stretches of 200-350 amino acids that exhibit a high sequence homology to the protein domains that were originally found in the Drosophila genes period (Per) and single-minded (Sim) and in AhR's dimerization partner the aryl hydrocarbon receptor nuclear translocator (ARNT).[7] The PAS domains support specific secondary interactions with other PAS domain containing proteins, as is the case with AhR and ARNT, so that dimeric and heteromeric protein complexes can form. The ligand binding site of AhR is contained within the PAS-B domain[8] and contains several conserved residues critical for ligand binding.[9] Finally, a glutamine-rich (Q-rich) domain is located in the C-terminal region of the protein and is involved in co-activator recruitment and transactivation.[10]
Ligands
AhR ligands have been generally classified into two categories, synthetic or naturally occurring. The first ligands to be discovered were synthetic and members of the halogenated aromatic hydrocarbons (polychlorinated dibenzodioxins, dibenzofurans and biphenyls) and polycyclic aromatic hydrocarbons (3-methylcholanthrene, benzo[a]pyrene, benzanthracenes and benzoflavones).[11][12] A range of synthetic ligands have been designed to target the possible future treatment of breast cancer.[13]
Research has focused on naturally occurring compounds with the hope of identifying an endogenous ligand. Naturally occurring compounds that have been identified as ligands of Ahr include derivatives of tryptophan such as indigo dye and indirubin,[14] tetrapyrroles such as bilirubin,[15] the arachidonic acid metabolites lipoxin A4 and prostaglandin G,[16] modified low-density lipoprotein[17] and several dietary carotenoids.[12] One assumption made in the search for an endogenous ligand is that the ligand will be a receptor agonist. However, work by Savouret et al. has shown this may not be the case since their findings demonstrate that 7-ketocholesterol competitively inhibits Ahr signal transduction.[18]
Carbidopa is a selective aryl hydrocarbon receptor modulator (SAhRM).[19] Other SAhRMs include microbial-derived 1,4-dihydroxy-2-napthoic acid[20] and plant-derived 3,3-diindolylmethane.[21]
Indolocarbazole (ICZ) is one of the strongest non-halogenated agonists for AhR in vitro reported.[22]
Ligand-independent AhR activity can be seen in mammalian AhR. The mammalian AhR needs no exogenous ligand-dependent activation to be functional, and this appears to be the case for its role in the regulation of the expression of some transforming growth factor-beta (TGF-b) isoforms. This is not to say that ligand-dependent AhR activation is not needed for the AhR to function in those cases, but that, if a ligand is needed, it is provided endogenously by the cells or tissues in question and its identity is unknown.[23]
Signaling pathway
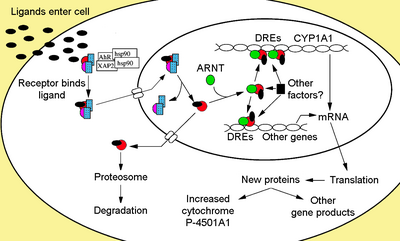

Cytosolic complex
Non-ligand bound AhR is retained in the cytoplasm as an inactive protein complex consisting of a dimer of Hsp90,[24][25] prostaglandin E synthase 3 (PTGES3, p23)[26][27][28][29] and a single molecule of the immunophilin-like AH receptor-interacting protein, also known as hepatitis B virus X-associated protein 2 (XAP2),[30] AhR interacting protein (AIP),[31][32] and AhR-activated 9 (ARA9).[33] The dimer of Hsp90, along with PTGES3 (p23), has a multifunctional role in the protection of the receptor from proteolysis, constraining the receptor in a conformation receptive to ligand binding and preventing the premature binding of ARNT.[8][27][29][34][35][36] AIP interacts with carboxyl-terminal of Hsp90 and binds to the AhR nuclear localization sequence (NLS) preventing the inappropriate trafficking of the receptor into the nucleus.[37][38][39]
Transforming growth factor-beta (TGF-β) signaling pathway
TGF-β cytokines are members of a signaling protein family that includes activin, Nodal subfamily, bone morphogenetic proteins, growth and differentiation factors, and Müllerian inhibitor subfamily. TGF-β signaling plays an important role in cell physiology and development by inhibiting cell proliferation, promoting apoptosis, inducing differentiation, and determining developmental fate in vertebrates and invertebrates.[40] TGF-β activators include proteases such as plasmin, cathepsins, and calpains. Thrombospondin 1, a glycoprotein that inhibits angiogenesis, and matrix metalloproteinase 2 (MMP-2). The extracellular matrix itself appears to play an important regulatory role in TGF-β signaling.[41][42]
Receptor activation
Upon ligand binding to AhR, AIP is released resulting in exposure of the NLS, which is located in the bHLH region,[43] leading to import into the nucleus.[44] It is presumed that once in the nucleus, Hsp90 dissociates exposing the two PAS domains allowing the binding of ARNT.[36][45][46][47] The activated AhR/ARNT heterodimer complex is then capable of either directly or indirectly interacting with DNA by binding to recognition sequences located in the 5’- regulatory region of dioxin-responsive genes.[36][46][48]
DNA binding (xenobiotic response element – XRE)
The classical recognition motif of the AhR/ARNT complex, referred to as either the AhR-, dioxin- or xenobiotic- responsive element (AHRE, DRE or XRE), contains the core sequence 5'-GCGTG-3'[49] within the consensus sequence 5'-T/GNGCGTGA/CG/CA-3'[50][51] in the promoter region of AhR responsive genes. The AhR/ARNT heterodimer directly binds the AHRE/DRE/XRE core sequence in an asymmetric manner such that ARNT binds to 5'-GTG-3' and AhR binding 5'-TC/TGC-3'.[52][53][54] Recent research suggests that a second type of element termed AHRE-II, 5'-CATG(N6)C[T/A]TG-3', is capable of indirectly acting with the AhR/ARNT complex.[55][56] Regardless of the response element, the result is a variety of differential changes in gene expression.
Functional role in physiology and toxicology
Role in development
In terms of evolution, the oldest physiological role of AhR is in development. AhR is presumed to have evolved from invertebrates where it served a ligand-independent role in normal development processes.[57] The AhR homolog in Drosophila, spineless (ss) is necessary for development of the distal segments of the antenna and leg.[58][59] Ss dimerizes with tango (tgo), which is the homolog to the mammalian Arnt, to initiate gene transcription. Evolution of the receptor in vertebrates resulted in the ability to bind ligands and might have helped humans evolve to tolerate smoky fires. In developing vertebrates, AhR seemingly plays a role in cellular proliferation and differentiation.[60] Despite lacking a clear endogenous ligand, AhR appears to play a role in the differentiation of many developmental pathways, including hematopoiesis,[61] lymphoid systems,[62][63] T-cells,[64] neurons,[65] and hepatocytes.[66] AhR has also been found to have an important function in hematopoietic stem cells: AhR antagonism promotes their self-renewal and ex-vivo expansion[67] and is involved in megakaryocyte differentiation.[68] In adulthood, signaling is associated with the stress response and mutations in AhR are associated with major depressive disorder.[69]
Adaptive and innate response
The adaptive response is manifested as the induction of xenobiotic metabolizing enzymes. Evidence of this response was first observed from the induction of cytochrome P450, family 1, subfamily A, polypeptide 1 (Cyp1a1) resultant from TCDD exposure, which was determined to be directly related to activation of the AhR signaling pathway.[70][71][72] The search for other metabolizing genes induced by AhR ligands, due to the presence of DREs, has led to the identification of an "AhR gene battery" of Phase I and Phase II metabolizing enzymes consisting of CYP1A1, CYP1A2, CYP1B1, NQO1, ALDH3A1, UGT1A2 and GSTA1.[73] Presumably, vertebrates have this function to be able to detect a wide range of chemicals, indicated by the wide range of substrates AhR is able to bind and facilitate their biotransformation and elimination. The AhR may also signal the presence of toxic chemicals in food and cause aversion of such foods.[74]
AhR activation seems to be also important for immunological responses and inhibiting inflammation[63] through upregulation of interleukin 22[75] and downregulation of Th17 response.[76] The Knockdown of AHR mostly downregulates the expression of innate immunity genes in THP-1 cells.[77]
Toxic response
Extensions of the adaptive response are the toxic responses elicited by AhR activation. Toxicity results from two different ways of AhR signaling. The first is a side effect of the adaptive response in which the induction of metabolizing enzymes results in the production of toxic metabolites. For example, the polycyclic aromatic hydrocarbon benzo[a]pyrene (BaP), a ligand for AhR, induces its own metabolism and bioactivation to a toxic metabolite via the induction of CYP1A1 and CYP1B1 in several tissues.[78] The second approach to toxicity is the result of aberrant changes in global gene transcription beyond those observed in the "AhR gene battery." These global changes in gene expression lead to adverse changes in cellular processes and function.[79] Microarray analysis has proved most beneficial in understanding and characterizing this response.[60][80][81][82]
Xenobiotic metabolizing enzymes help with the metabolic process by transforming and the excretion of chemicals. The most potent inducer of CYP1A1 is 2,3,7,8-tetrachlorodibenzo-p-dioxin (TCDD). In addition, TCDD induces a broad spectrum of biochemical and toxic effects, such as teratogenesis, immunosuppression and tumor promotion. Most, if not all, of the effects caused by TCDD and other PAHs are known to be mediated by AhR which has a high binding affinity to TCDD.[40]
Protein-protein interactions
In addition to the protein interactions mentioned above, AhR has also been shown to interact with the following:
References
- ↑ "The Aryl Hydrocarbon Receptor in Immunity: Tools and Potential". Suppression and Regulation of Immune Responses. Methods in Molecular Biology. 1371. 2016. pp. 239–57. doi:10.1007/978-1-4939-3139-2_16. ISBN 978-1-4939-3138-5.
- ↑ "The aryl hydrocarbon receptor: a multifunctional chemical sensor for host defense and homeostatic maintenance". Experimental Animals 66 (2): 75–89. May 2017. doi:10.1538/expanim.16-0092. PMID 27980293.
- ↑ "Regulation of the Immune Response by the Aryl Hydrocarbon Receptor". Immunity 48 (1): 19–33. January 2018. doi:10.1016/j.immuni.2017.12.012. PMID 29343438.
- ↑ "Cloning of the Ah-receptor cDNA reveals a distinctive ligand-activated transcription factor". Proceedings of the National Academy of Sciences of the United States of America 89 (17): 8185–8189. September 1992. doi:10.1073/pnas.89.17.8185. PMID 1325649. Bibcode: 1992PNAS...89.8185B.
- ↑ "Identification of functional domains of the aryl hydrocarbon receptor". The Journal of Biological Chemistry 270 (49): 29270–29278. December 1995. doi:10.1074/jbc.270.49.29270. PMID 7493958.
- ↑ "An overview of the basic helix-loop-helix proteins". Genome Biology 5 (6): 226. 2004. doi:10.1186/gb-2004-5-6-226. PMID 15186484.
- ↑ "cDNA cloning and structure of mouse putative Ah receptor". Biochemical and Biophysical Research Communications 184 (1): 246–253. April 1992. doi:10.1016/0006-291X(92)91185-S. PMID 1314586.
- ↑ 8.0 8.1 "Definition of a minimal domain of the dioxin receptor that is associated with Hsp90 and maintains wild type ligand binding affinity and specificity". The Journal of Biological Chemistry 270 (42): 25291–25300. October 1995. doi:10.1074/jbc.270.42.25291. PMID 7559670.
- ↑ "Identification of amino acid residues in the Ah receptor involved in ligand binding". Biochemical and Biophysical Research Communications 354 (2): 396–402. March 2007. doi:10.1016/j.bbrc.2006.12.227. PMID 17227672.
- ↑ "The Q-rich subdomain of the human Ah receptor transactivation domain is required for dioxin-mediated transcriptional activity". The Journal of Biological Chemistry 276 (45): 42302–42310. November 2001. doi:10.1074/jbc.M104798200. PMID 11551916.
- ↑ "Ligand binding and activation of the Ah receptor". Chemico-Biological Interactions 141 (1–2): 3–24. September 2002. doi:10.1016/S0009-2797(02)00063-7. PMID 12213382. https://escholarship.org/uc/item/1c68w9nb.
- ↑ 12.0 12.1 12.2 "Activation of the aryl hydrocarbon receptor by structurally diverse exogenous and endogenous chemicals". Annual Review of Pharmacology and Toxicology 43: 309–334. 2003. doi:10.1146/annurev.pharmtox.43.100901.135828. PMID 12540743.
- ↑ "The aryl hydrocarbon receptor (AhR) as a breast cancer drug target". Medicinal Research Reviews 40 (3): 972–1001. May 2020. doi:10.1002/med.21645. PMID 31721255.
- ↑ "Indirubin and indigo are potent aryl hydrocarbon receptor ligands present in human urine". The Journal of Biological Chemistry 276 (34): 31475–31478. August 2001. doi:10.1074/jbc.C100238200. PMID 11425848.
- ↑ "Aryl hydrocarbon receptor-dependent induction of cyp1a1 by bilirubin in mouse hepatoma hepa 1c1c7 cells". Molecular Pharmacology 52 (4): 590–599. October 1997. doi:10.1124/mol.52.4.590. PMID 9380021.
- ↑ "Activation of the Ah receptor signaling pathway by prostaglandins". Journal of Biochemical and Molecular Toxicology 15 (4): 187–196. 2001. doi:10.1002/jbt.16. PMID 11673847.
- ↑ "The aryl hydrocarbon receptor is activated by modified low-density lipoprotein". Proceedings of the National Academy of Sciences of the United States of America 104 (4): 1412–1417. January 2007. doi:10.1073/pnas.0607296104. PMID 17227852. Bibcode: 2007PNAS..104.1412M.
- ↑ "7-ketocholesterol is an endogenous modulator for the arylhydrocarbon receptor". The Journal of Biological Chemistry 276 (5): 3054–3059. February 2001. doi:10.1074/jbc.M005988200. PMID 11042205.
- ↑ "Carbidopa: a selective Ah receptor modulator (SAhRM)". The Biochemical Journal 474 (22): 3763–3765. November 2017. doi:10.1042/BCJ20170728. PMID 29109131.
- ↑ "Editor's Highlight: Microbial-Derived 1,4-Dihydroxy-2-naphthoic Acid and Related Compounds as Aryl Hydrocarbon Receptor Agonists/Antagonists: Structure-Activity Relationships and Receptor Modeling". Toxicological Sciences 155 (2): 458–473. February 2017. doi:10.1093/toxsci/kfw230. PMID 27837168.
- ↑ "A selective aryl hydrocarbon receptor modulator 3,3'-Diindolylmethane inhibits gastric cancer cell growth". Journal of Experimental & Clinical Cancer Research 31 (1): 46. May 2012. doi:10.1186/1756-9966-31-46. PMID 22592002.
- ↑ "Synthesis and biological evaluation of fused thio- and selenopyrans as new indolocarbazole analogues with aryl hydrocarbon receptor affinity". Bioorganic & Medicinal Chemistry 17 (4): 1648–1653. February 2009. doi:10.1016/j.bmc.2008.12.072. PMID 19186062.
- ↑ "Ligand binding and activation of the Ah receptor". Chemico-Biological Interactions 141 (1–2): 3–24. September 2002. doi:10.1016/S0009-2797(02)00063-7. PMID 12213382. https://escholarship.org/uc/item/1c68w9nb.
- ↑ "Association of the dioxin receptor with the Mr 90,000 heat shock protein: a structural kinship with the glucocorticoid receptor". Biochemical and Biophysical Research Communications 155 (2): 801–807. September 1988. doi:10.1016/S0006-291X(88)80566-7. PMID 2844180.
- ↑ "Association of the Ah receptor with the 90-kDa heat shock protein". The Journal of Biological Chemistry 263 (27): 13802–13805. September 1988. doi:10.1016/S0021-9258(18)68314-0. PMID 2843537.
- ↑ "Cooperation of heat shock protein 90 and p23 in aryl hydrocarbon receptor signaling". Cell Stress & Chaperones 9 (1): 4–20. March 2004. doi:10.1379/460.1. PMID 15270073.
- ↑ 27.0 27.1 "Evidence that the co-chaperone p23 regulates ligand responsiveness of the dioxin (Aryl hydrocarbon) receptor". The Journal of Biological Chemistry 274 (19): 13519–13524. May 1999. doi:10.1074/jbc.274.19.13519. PMID 10224120.
- ↑ "The hsp90 chaperone complex regulates intracellular localization of the dioxin receptor". Molecular and Cellular Biology 21 (7): 2594–2607. April 2001. doi:10.1128/MCB.21.7.2594-2607.2001. PMID 11259606.
- ↑ 29.0 29.1 "P23 enhances the formation of the aryl hydrocarbon receptor-DNA complex". Biochemical Pharmacology 65 (6): 941–948. March 2003. doi:10.1016/S0006-2952(02)01650-7. PMID 12623125.
- ↑ "Hepatitis B virus X-associated protein 2 is a subunit of the unliganded aryl hydrocarbon receptor core complex and exhibits transcriptional enhancer activity". Molecular and Cellular Biology 18 (2): 978–988. February 1998. doi:10.1128/MCB.18.2.978. PMID 9447995.
- ↑ "A novel cytoplasmic protein that interacts with the Ah receptor, contains tetratricopeptide repeat motifs, and augments the transcriptional response to 2,3,7,8-tetrachlorodibenzo-p-dioxin". The Journal of Biological Chemistry 272 (14): 8878–8884. April 1997. doi:10.1074/jbc.272.14.8878. PMID 9083006.
- ↑ 32.0 32.1 "Aryl Hydrocarbon Receptor Interacting Protein Targets IRF7 to Suppress Antiviral Signaling and the Induction of Type I Interferon". The Journal of Biological Chemistry 290 (23): 14729–14739. June 2015. doi:10.1074/jbc.M114.633065. PMID 25911105.
- ↑ "Ligand-dependent interaction of the aryl hydrocarbon receptor with a novel immunophilin homolog in vivo". The Journal of Biological Chemistry 272 (17): 11452–11456. April 1997. doi:10.1074/jbc.272.17.11452. PMID 9111057.
- ↑ "The 90-kDa heat shock protein is essential for Ah receptor signaling in a yeast expression system". The Journal of Biological Chemistry 269 (48): 30109–30112. December 1994. doi:10.1016/S0021-9258(18)43782-9. PMID 7982913.
- ↑ "Dual roles of the 90-kDa heat shock protein hsp90 in modulating functional activities of the dioxin receptor. Evidence that the dioxin receptor functionally belongs to a subclass of nuclear receptors which require hsp90 both for ligand binding activity and repression of intrinsic DNA binding activity". The Journal of Biological Chemistry 267 (19): 13728–13734. July 1992. doi:10.1016/S0021-9258(18)42274-0. PMID 1320028.
- ↑ 36.0 36.1 36.2 "Ligand-dependent recruitment of the Arnt coregulator determines DNA recognition by the dioxin receptor". Molecular and Cellular Biology 13 (4): 2504–2514. April 1993. doi:10.1128/MCB.13.4.2504. PMID 8384309.
- ↑ "Characterization of the Ah receptor-associated protein, ARA9". The Journal of Biological Chemistry 273 (50): 33580–33587. December 1998. doi:10.1074/jbc.273.50.33580. PMID 9837941.
- ↑ "Subcellular localization of the aryl hydrocarbon receptor is modulated by the immunophilin homolog hepatitis B virus X-associated protein 2". The Journal of Biological Chemistry 275 (48): 37448–37453. December 2000. doi:10.1074/jbc.M006873200. PMID 10986286.
- ↑ "The hsp90 Co-chaperone XAP2 alters importin beta recognition of the bipartite nuclear localization signal of the Ah receptor and represses transcriptional activity". The Journal of Biological Chemistry 278 (4): 2677–2685. January 2003. doi:10.1074/jbc.M209331200. PMID 12431985.
- ↑ 40.0 40.1 "Functional role of AhR in the expression of toxic effects by TCDD". Biochimica et Biophysica Acta (BBA) - General Subjects. Cellular Biology of Cytochrome P450 Regulation 1619 (3): 263–268. February 2003. doi:10.1016/S0304-4165(02)00485-3. PMID 12573486.
- ↑ "Ah receptor signals cross-talk with multiple developmental pathways". Biochemical Pharmacology 69 (2): 199–207. January 2005. doi:10.1016/j.bcp.2004.06.043. PMID 15627472.
- ↑ "The aryl hydrocarbon receptor cross-talks with multiple signal transduction pathways". Biochemical Pharmacology 77 (4): 713–722. February 2009. doi:10.1016/j.bcp.2008.08.031. PMID 18817753.
- ↑ "Nuclear localization and export signals of the human aryl hydrocarbon receptor". The Journal of Biological Chemistry 273 (5): 2895–2904. January 1998. doi:10.1074/jbc.273.5.2895. PMID 9446600.
- ↑ "Analysis of the complex relationship between nuclear export and aryl hydrocarbon receptor-mediated gene regulation". Molecular and Cellular Biology 20 (16): 6095–6104. August 2000. doi:10.1128/MCB.20.16.6095-6104.2000. PMID 10913191.
- ↑ "Cloning of a factor required for activity of the Ah (dioxin) receptor". Science 252 (5008): 954–958. May 1991. doi:10.1126/science.1852076. PMID 1852076. Bibcode: 1991Sci...252..954H.
- ↑ 46.0 46.1 "Role of the aryl hydrocarbon receptor nuclear translocator protein in aryl hydrocarbon (dioxin) receptor action". Molecular Pharmacology 44 (3): 511–518. September 1993. PMID 8396713.
- ↑ "Identification of the Ah receptor nuclear translocator protein (Arnt) as a component of the DNA binding form of the Ah receptor". Science 256 (5060): 1193–1195. May 1992. doi:10.1126/science.256.5060.1193. PMID 1317062. Bibcode: 1992Sci...256.1193R.
- ↑ "In vitro analysis of Ah receptor domains involved in ligand-activated DNA recognition". Proceedings of the National Academy of Sciences of the United States of America 90 (18): 8566–8570. September 1993. doi:10.1073/pnas.90.18.8566. PMID 8397410. Bibcode: 1993PNAS...90.8566D.
- ↑ "Protein-DNA interactions at a dioxin-responsive enhancer. Mutational analysis of the DNA-binding site for the liganded Ah receptor". The Journal of Biological Chemistry 267 (10): 6815–6819. April 1992. doi:10.1016/S0021-9258(19)50499-9. PMID 1313023.
- ↑ "Protein-DNA interactions at a dioxin-responsive enhancer. Analysis of six bona fide DNA-binding sites for the liganded Ah receptor". The Journal of Biological Chemistry 268 (9): 6575–6580. March 1993. doi:10.1016/S0021-9258(18)53289-0. PMID 8384216.
- ↑ "DNA sequence determinants for binding of transformed Ah receptor to a dioxin-responsive enhancer". Biochemistry 31 (21): 5060–5067. June 1992. doi:10.1021/bi00136a019. PMID 1318077.
- ↑ "Control of CNS midline transcription by asymmetric E-box-like elements: similarity to xenobiotic responsive regulation". Development 120 (12): 3563–3569. December 1994. doi:10.1242/dev.120.12.3563. PMID 7821222.
- ↑ "Orientation of the heterodimeric aryl hydrocarbon (dioxin) receptor complex on its asymmetric DNA recognition sequence". Molecular Pharmacology 47 (3): 432–438. March 1995. PMID 7700240.
- ↑ "DNA binding specificities and pairing rules of the Ah receptor, ARNT, and SIM proteins". The Journal of Biological Chemistry 270 (44): 26292–26302. November 1995. doi:10.1074/jbc.270.44.26292. PMID 7592839.
- ↑ "Dioxin-responsive AHRE-II gene battery: identification by phylogenetic footprinting". Biochemical and Biophysical Research Communications 321 (3): 707–715. August 2004. doi:10.1016/j.bbrc.2004.06.177. PMID 15358164.
- ↑ "A novel induction mechanism of the rat CYP1A2 gene mediated by Ah receptor-Arnt heterodimer". Biochemical and Biophysical Research Communications 318 (3): 746–755. June 2004. doi:10.1016/j.bbrc.2004.04.090. PMID 15144902.
- ↑ "Unexpected diversity of aryl hydrocarbon receptors in non-mammalian vertebrates: insights from comparative genomics". Journal of Experimental Zoology Part A: Comparative Experimental Biology 305 (9): 693–706. September 2006. doi:10.1002/jez.a.323. PMID 16902966.
- ↑ "Control of distal antennal identity and tarsal development in Drosophila by spineless-aristapedia, a homolog of the mammalian dioxin receptor". Genes & Development 12 (9): 1290–1303. May 1998. doi:10.1101/gad.12.9.1290. PMID 9573046.
- ↑ "The spineless-aristapedia and tango bHLH-PAS proteins interact to control antennal and tarsal development in Drosophila". Development 126 (17): 3937–3945. September 1999. doi:10.1242/dev.126.17.3937. PMID 10433921. https://cdr.lib.unc.edu/downloads/b8515x68n.
- ↑ 60.0 60.1 "Aryl hydrocarbon receptor regulates distinct dioxin-dependent and dioxin-independent gene batteries". Molecular Pharmacology 69 (1): 140–153. January 2006. doi:10.1124/mol.105.018705. PMID 16214954.
- ↑ "The aryl hydrocarbon receptor has an important role in the regulation of hematopoiesis: implications for benzene-induced hematopoietic toxicity". Chemico-Biological Interactions 184 (1–2): 246–251. March 2010. doi:10.1016/j.cbi.2009.10.019. PMID 19896476.
- ↑ "Natural aryl hydrocarbon receptor ligands control organogenesis of intestinal lymphoid follicles". Science 334 (6062): 1561–1565. December 2011. doi:10.1126/science.1214914. PMID 22033518. Bibcode: 2011Sci...334.1561K.
- ↑ 63.0 63.1 "Exogenous stimuli maintain intraepithelial lymphocytes via aryl hydrocarbon receptor activation". Cell 147 (3): 629–640. October 2011. doi:10.1016/j.cell.2011.09.025. PMID 21999944.
- ↑ "Control of T(reg) and T(H)17 cell differentiation by the aryl hydrocarbon receptor". Nature 453 (7191): 65–71. May 2008. doi:10.1038/nature06880. PMID 18362915.
- ↑ "Over-expression of AhR (aryl hydrocarbon receptor) induces neural differentiation of Neuro2a cells: neurotoxicology study". Environmental Health 5: 24. September 2006. doi:10.1186/1476-069X-5-24. PMID 16956419.
- ↑ "Aryl hydrocarbon receptor-dependent liver development and hepatotoxicity are mediated by different cell types". Proceedings of the National Academy of Sciences of the United States of America 102 (49): 17858–17863. December 2005. doi:10.1073/pnas.0504757102. PMID 16301529. Bibcode: 2005PNAS..10217858W.
- ↑ "Aryl hydrocarbon receptor antagonists promote the expansion of human hematopoietic stem cells". Science 329 (5997): 1345–1348. September 2010. doi:10.1126/science.1191536. PMID 20688981. Bibcode: 2010Sci...329.1345B.
- ↑ "The aryl hydrocarbon receptor (AHR) transcription factor regulates megakaryocytic polyploidization". British Journal of Haematology 152 (4): 469–484. February 2011. doi:10.1111/j.1365-2141.2010.08548.x. PMID 21226706.
- ↑ "Microbial metabolites and immune regulation: New targets for major depressive disorder". Brain, Behavior, & Immunity - Health 9: 100169. December 2020. doi:10.1016/j.bbih.2020.100169. PMID 34589904.
- ↑ "Induction of mRNA specific for cytochrome P1-450 in wild type and variant mouse hepatoma cells". The Journal of Biological Chemistry 258 (17): 10390–10394. September 1983. doi:10.1016/S0021-9258(17)44469-3. PMID 6885786.
- ↑ "Regulation of cytochrome P1-450 gene transcription by 2,3,7, 8-tetrachlorodibenzo-p-dioxin in wild type and variant mouse hepatoma cells". The Journal of Biological Chemistry 259 (9): 5400–5402. May 1984. doi:10.1016/S0021-9258(18)91022-7. PMID 6715350.
- ↑ "Dioxin-induced CYP1A1 transcription in vivo: the aromatic hydrocarbon receptor mediates transactivation, enhancer-promoter communication, and changes in chromatin structure". Molecular and Cellular Biology 16 (1): 430–436. January 1996. doi:10.1128/MCB.16.1.430. PMID 8524325.
- ↑ "Role of the aromatic hydrocarbon receptor and [Ah] gene battery in the oxidative stress response, cell cycle control, and apoptosis". Biochemical Pharmacology 59 (1): 65–85. January 2000. doi:10.1016/S0006-2952(99)00310-X. PMID 10605936.
- ↑ "Immediate and highly sensitive aversion response to a novel food item linked to AH receptor stimulation". Toxicology Letters 203 (3): 252–257. June 2011. doi:10.1016/j.toxlet.2011.03.025. PMID 21458548.
- ↑ "Aryl hydrocarbon receptor-induced signals up-regulate IL-22 production and inhibit inflammation in the gastrointestinal tract". Gastroenterology 141 (1): 237–48, 248.e1. July 2011. doi:10.1053/j.gastro.2011.04.007. PMID 21600206.
- ↑ "An aryl hydrocarbon receptor ligand acts on dendritic cells and T cells to suppress the Th17 response in allergic rhinitis patients". Laboratory Investigation; A Journal of Technical Methods and Pathology 94 (5): 528–535. May 2014. doi:10.1038/labinvest.2014.8. PMID 24514067.
- ↑ "Engagement of the Aryl Hydrocarbon Receptor in Mycobacterium tuberculosis-Infected Macrophages Has Pleiotropic Effects on Innate Immune Signaling". Journal of Immunology 195 (9): 4479–4491. November 2015. doi:10.4049/jimmunol.1501141. PMID 26416282.
- ↑ "DNA adduct formation in precision-cut rat liver and lung slices exposed to benzo[a]pyrene". Toxicological Sciences 77 (2): 307–314. February 2004. doi:10.1093/toxsci/kfh030. PMID 14691214.
- ↑ "Dioxins, the aryl hydrocarbon receptor and the central regulation of energy balance". Frontiers in Neuroendocrinology 31 (4): 452–478. October 2010. doi:10.1016/j.yfrne.2010.07.002. PMID 20624415.
- ↑ "Differential toxicogenomic responses to 2,3,7,8-tetrachlorodibenzo-p-dioxin in malignant and nonmalignant human airway epithelial cells". Toxicological Sciences 69 (2): 409–423. October 2002. doi:10.1093/toxsci/69.2.409. PMID 12377990.
- ↑ "Subchronic exposure to TCDD, PeCDF, PCB126, and PCB153: effect on hepatic gene expression". Environmental Health Perspectives 112 (16): 1636–1644. November 2004. doi:10.1289/ehp.7253. PMID 15598615.
- ↑ "Hepatic gene downregulation following acute and subchronic exposure to 2,3,7,8-tetrachlorodibenzo-p-dioxin". Toxicological Sciences 94 (2): 428–438. December 2006. doi:10.1093/toxsci/kfl111. PMID 16984957.
- ↑ "Characterization of a subset of the basic-helix-loop-helix-PAS superfamily that interacts with components of the dioxin signaling pathway". The Journal of Biological Chemistry 272 (13): 8581–8593. March 1997. doi:10.1074/jbc.272.13.8581. PMID 9079689.
- ↑ "Interactions between the aryl hydrocarbon receptor and P-TEFb. Sequential recruitment of transcription factors and differential phosphorylation of C-terminal domain of RNA polymerase II at cyp1a1 promoter". The Journal of Biological Chemistry 278 (45): 44041–44048. November 2003. doi:10.1074/jbc.M306443200. PMID 12917420.
- ↑ "The aryl hydrocarbon receptor mediates degradation of estrogen receptor alpha through activation of proteasomes". Molecular and Cellular Biology 23 (6): 1843–1855. March 2003. doi:10.1128/MCB.23.6.1843-1855.2003. PMID 12612060.
- ↑ "The aryl hydrocarbon receptor interacts with estrogen receptor alpha and orphan receptors COUP-TFI and ERRalpha1". Archives of Biochemistry and Biophysics 373 (1): 163–174. January 2000. doi:10.1006/abbi.1999.1552. PMID 10620335.
- ↑ "Recruitment of the NCoA/SRC-1/p160 family of transcriptional coactivators by the aryl hydrocarbon receptor/aryl hydrocarbon receptor nuclear translocator complex". Molecular and Cellular Biology 22 (12): 4319–4333. June 2002. doi:10.1128/MCB.22.12.4319-4333.2002. PMID 12024042.
- ↑ "Interaction with Nedd8, a ubiquitin-like protein, enhances the transcriptional activity of the aryl hydrocarbon receptor". The Journal of Biological Chemistry 277 (46): 44028–44034. November 2002. doi:10.1074/jbc.M202413200. PMID 12215427.
- ↑ "Differential recruitment of coactivator RIP140 by Ah and estrogen receptors. Absence of a role for LXXLL motifs". The Journal of Biological Chemistry 274 (32): 22155–22164. August 1999. doi:10.1074/jbc.274.32.22155. PMID 10428779.
- ↑ "The RelA NF-kappaB subunit and the aryl hydrocarbon receptor (AhR) cooperate to transactivate the c-myc promoter in mammary cells". Oncogene 19 (48): 5498–5506. November 2000. doi:10.1038/sj.onc.1203945. PMID 11114727.
- ↑ "2,3,7,8-Tetrachlorodibenzo-p-dioxin suppresses tumor necrosis factor-alpha and anti-CD40-induced activation of NF-kappaB/Rel in dendritic cells: p50 homodimer activation is not affected". Molecular Pharmacology 62 (3): 722–728. September 2002. doi:10.1124/mol.62.3.722. PMID 12181450.
- ↑ "RelB, a new partner of aryl hydrocarbon receptor-mediated transcription". Molecular Endocrinology 21 (12): 2941–2955. December 2007. doi:10.1210/me.2007-0211. PMID 17823304.
- ↑ "A direct interaction between the aryl hydrocarbon receptor and retinoblastoma protein. Linking dioxin signaling to the cell cycle". The Journal of Biological Chemistry 273 (35): 22708–22713. August 1998. doi:10.1074/jbc.273.35.22708. PMID 9712901.
External links
- Aryl+hydrocarbon+receptor at the US National Library of Medicine Medical Subject Headings (MeSH)
- Human AHR genome location and AHR gene details page in the UCSC Genome Browser.
- Human ARNT genome location and ARNT gene details page in the UCSC Genome Browser.
 |

