Biology:NFAT5
 Generic protein structure example |
Nuclear factor of activated T-cells 5, also known as NFAT5 and sometimes TonEBP, is a human gene that encodes a transcription factor that regulates the expression of genes involved in the osmotic stress.[1]
The product of this gene is a member of the nuclear factors of activated T cells (NFAT) family of transcription factors. Proteins belonging to this family play a central role in inducible gene transcription during the immune response. This protein regulates gene expression induced by osmotic stress in mammalian cells. Unlike monomeric members of this protein family, this protein exists as a homodimer and forms stable dimers with DNA elements. Multiple transcript variants encoding different isoforms have been found for this gene.[1]
Osmotic stress
Tissues that comprise the kidneys, skin, and eyes are often subjected to osmotic stresses. When the extracellular environment is hypertonic, cells lose water and consequently, shrink. To counteract this, cells increase their sodium uptake in order to lose less water. However, an increase in intracellular ionic concentration is harmful to the cell. Cells can alternatively synthesize enzymes and transporters that increase intracellular concentration of organic osmolytes, which are less toxic than excess ions but which also aid in water retention. Under conditions of hyperosmolarity, NFAT5 is synthesized and accumulates in the nucleus. NFAT5 stimulates the transcription of genes for aldose reductase (AR), the sodium chloride-betaine cotransporter (SLC6A12) the sodium/myo-inositol cotransporter (SLC5A3), the taurine transporter (SLC6A6) and neuropathy target esterase which are involved in the production and uptake of organic osmolytes.[2][3] Additionally, NFAT5 induces heat shock proteins, Hsp70, and osmotic stress proteins. NFAT5 is also implicated in cytokine production.[4]
It has been shown that when NFAT5 is inhibited in renal and immune cells, these cells become significantly more susceptible to osmotic stress. NFAT5 deficient mice were found to suffer from massive cell loss in the renal medulla.[5] Additionally, mice expressing a dominant-negative form of NFAT5 in their eyes exhibited decreased viability under hypertonic extracellular environment.[6]
Structure
The NFAT family consists of five different forms: NFAT1, NFAT2, NFAT3, NFAT4, and NFAT5 (this protein). The proteins in this family are expressed in nearly every tissue in the body and are known transcriptional regulators in cytokine and immune cell expression. Among the different forms of NFAT, NFAT5 is an important component of the hyperosmolar stress response system.[4] cDNA of NFAT5 was first isolated from a human brain cDNA library. Subsequent analysis revealed that NFAT5 is a member of the Rel family, which also consists of NF-κB and NFATc proteins. The largest Rel protein, it consists of nearly 1,500 amino acid residues. Like the other Rel proteins, NFAT5 contains the Rel homology domain, a conserved DNA-binding domain. Outside of the Rel homology domain, no similarities exist between NFAT5 and NF-κB or NFATc. Among these differences is the absence of docking sites for calcineurin, which is necessary for NFATc nuclear import.[7] Instead, NFAT5 is a constitutively nuclear protein whose activity and localization does not depend on calcineurin-mediated dephosphorylation.[4][7] Increased NFAT5 transcription is correlated with p38 MAPK-mediated phosphorylation.
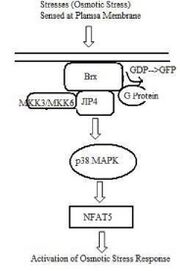
Mechanism of Activation
Although the precise mechanism by which osmotic stress is sensed by the cell is unclear, it has been suggested that Brx, a guanine nucleotide exchange factor (GEF) localized near the plasma membrane, is activated by osmotic stress through changes in the cytoskeleton structure. Alternatively, Brx may also be activated through changes in its interactions with possible osmosensor molecules at the cell membrane.[8] Upon Brx activation, the GEF domain of Brx facilitates activation of Rho-type small G proteins from its inactive GDP state to active GTP state. Additionally, activated Brx also recruits and physically interacts with JIP4, a p38 MAPK-specific scaffold protein. JIP4 binds to downstream kinases, MKK3 and MKK6.[9] This complex then activates p38 mitogen-activated protein kinase (MAPK). Activation of p38 MAPK is regulated by Cdc42 and Rac1. Activation of p38 MAPK is a necessary step for NFAT5 expression.[8]
It has been found that NFAT5 expression, following hyperosmolarity, depends on p38 mitogen-activated protein kinase (MAPK). The addition of a p38 MAPK inhibitor was found to correlate with decreased NFAT5 expression, even in the presence of osmotic stress signals.[5] However, the downstream transcription of the NFAT5 gene by p38 MAPK is currently not yet characterized. It is hypothesized that p38 MAPK phosphorylation activates c-Fos and interferon regulatory factors (IRFs), which bind to AP-1-binding sites and ISRES (Interferon Stimulated Response Element) respectively. Binding to these sites consequently activates the transcription of target genes.[8]
Although the Brx-mediated activation of NFAT5 has only been examined in lymphocyte response to osmotic stress, it is hypothesized that this mechanism is a common one in other cell types.
Additional Roles
NFAT5 has also been implicated in other biological roles, such as in embryonic development. Mice in the embryonic stages with non-function NFAT5 exhibited reduced survivorship.
NFAT5 is also involved in cellular proliferation. NFAT5 mRNA expression is particularly high in proliferating cells. Inhibition of NFAT5 in embryonic fibroblasts resulted in cell cycle arrest.[4]
Although NFAT5 has been found to be important in other biological processes besides hyperosmotic stress response, the mechanism by which NFAT5 acts in these other processes are currently not well known.
References
- ↑ 1.0 1.1 "Entrez Gene: NFAT5 nuclear factor of activated T-cells 5, tonicity-responsive". https://www.ncbi.nlm.nih.gov/sites/entrez?Db=gene&Cmd=ShowDetailView&TermToSearch=10725.
- ↑ "TonEBP stimulates multiple cellular pathways for adaptation to hypertonic stress: Organic osmolyte-dependent and -independent pathways". AJP: Renal Physiology 300 (3): F707–F715. 2011. doi:10.1152/ajprenal.00227.2010. PMID 21209002.
- ↑ "Tonicity-responsive enhancer binding protein, a Rel-like protein that stimulates transcription in response to hypertonicity". Proc Natl Acad Sci USA 96 (5): 2538–2542. 1999. doi:10.1073/pnas.96.5.2538. PMID 10051678. Bibcode: 1999PNAS...96.2538M.
- ↑ 4.0 4.1 4.2 4.3 "NFAT5 induction and its role in hyperosmolar stressed human limbal epithelial cells". Invest. Ophthalmol. Vis. Sci. 49 (5): 1827–1835. 2008. doi:10.1167/iovs.07-1142. PMID 18436816.
- ↑ 5.0 5.1 "Loss of NFAT5 results in renal atrophy and lack of tonicity-responsive gene expression". Proc. Natl. Acad. Sci. U.S.A. 101 (8): 2392–7. February 2004. doi:10.1073/pnas.0308703100. PMID 14983020. Bibcode: 2004PNAS..101.2392L.
- ↑ "Transgenic mice expressing dominant-negative osmotic-response element-binding protein (OREBP) in lens exhibit fiber cell elongation defect associated with increased DNA breaks". J. Biol. Chem. 280 (20): 19986–91. May 2005. doi:10.1074/jbc.M501689200. PMID 15774462.
- ↑ 7.0 7.1 "NFAT5, a constitutively nuclear NFAT protein that does not cooperate with Fos and Jun". Proc. Natl. Acad. Sci. U.S.A. 96 (13): 7214–9. June 1999. doi:10.1073/pnas.96.13.7214. PMID 10377394. Bibcode: 1999PNAS...96.7214L.
- ↑ 8.0 8.1 8.2 "Brx mediates the response of lymphocytes to osmotic stress through the activation of NFAT5". Sci Signal 2 (57): ra5. 2009. doi:10.1126/scisignal.2000081. PMID 19211510.
- ↑ "Role of the JIP4 scaffold protein in the regulation of mitogen-activated protein kinase signaling pathways". Mol. Cell. Biol. 25 (7): 2733–43. April 2005. doi:10.1128/MCB.25.7.2733-2743.2005. PMID 15767678.
Further reading
- "NF-AT5: the NF-AT family of transcription factors expands in a new direction.". Cold Spring Harb. Symp. Quant. Biol. 64 (1): 517–26. 2001. doi:10.1101/sqb.1999.64.517. PMID 11233530.
- "NFAT: ubiquitous regulator of cell differentiation and adaptation.". J. Cell Biol. 156 (5): 771–4. 2002. doi:10.1083/jcb.200111073. PMID 11877454.
- "Prediction of the coding sequences of unidentified human genes. XII. The complete sequences of 100 new cDNA clones from brain which code for large proteins in vitro.". DNA Res. 5 (6): 355–64. 1999. doi:10.1093/dnares/5.6.355. PMID 10048485.
- "Tonicity-responsive enhancer binding protein, a rel-like protein that stimulates transcription in response to hypertonicity.". Proc. Natl. Acad. Sci. U.S.A. 96 (5): 2538–42. 1999. doi:10.1073/pnas.96.5.2538. PMID 10051678. Bibcode: 1999PNAS...96.2538M.
- "NFAT5, a constitutively nuclear NFAT protein that does not cooperate with Fos and Jun.". Proc. Natl. Acad. Sci. U.S.A. 96 (13): 7214–9. 1999. doi:10.1073/pnas.96.13.7214. PMID 10377394. Bibcode: 1999PNAS...96.7214L.
- "Isolation and characterization of novel CAG repeat containing genes expressed in human brain.". DNA Seq. 10 (1): 1–6. 2000. doi:10.3109/10425179909033929. PMID 10565538.
- "Purification, identification, and characterization of an osmotic response element binding protein.". Biochem. Biophys. Res. Commun. 270 (1): 52–61. 2000. doi:10.1006/bbrc.2000.2376. PMID 10733904.
- "The NFAT-related protein NFATL1 (TonEBP/NFAT5) is induced upon T cell activation in a calcineurin-dependent manner.". J. Immunol. 165 (9): 4884–94. 2000. doi:10.4049/jimmunol.165.9.4884. PMID 11046013.
- "Assignment of transcription factor NFAT5 to human chromosome 16q22.1, murine chromosome 8D and porcine chromosome 6p1.4 and comparison of the polyglutamine domains.". Cytogenet. Cell Genet. 90 (1–2): 68–70. 2000. doi:10.1159/000015665. PMID 11060450.
- "Bridging the NFAT and NF-kappaB families: NFAT5 dimerization regulates cytokine gene transcription in response to osmotic stress.". Immunity 15 (1): 47–58. 2001. doi:10.1016/S1074-7613(01)00165-0. PMID 11485737.
- "Genomic organization of the human NFAT5 gene: exon-intron structure of the 14-kb transcript and CpG-island analysis of the promoter region.". Cytogenet. Cell Genet. 93 (3–4): 239–41. 2001. doi:10.1159/000056990. PMID 11528118.
- "Structure of a TonEBP-DNA complex reveals DNA encircled by a transcription factor.". Nat. Struct. Biol. 9 (2): 90–4. 2002. doi:10.1038/nsb749. PMID 11780147.
- "Activity of the TonEBP/OREBP transactivation domain varies directly with extracellular NaCl concentration.". Proc. Natl. Acad. Sci. U.S.A. 99 (2): 739–44. 2002. doi:10.1073/pnas.241637298. PMID 11792870. Bibcode: 2002PNAS...99..739F.
- "The characterization and purification of a human transcription factor modulating the glutathione peroxidase gene in response to oxygen tension.". Mol. Cell. Biochem. 229 (1–2): 73–83. 2002. doi:10.1023/A:1017921110363. PMID 11936849.
- "Fyn and p38 signaling are both required for maximal hypertonic activation of the osmotic response element-binding protein/tonicity-responsive enhancer-binding protein (OREBP/TonEBP).". J. Biol. Chem. 277 (48): 46085–92. 2003. doi:10.1074/jbc.M208138200. PMID 12359721.
- "The CD23b promoter is a target for NF-AT transcription factors in B-CLL cells.". Biochim. Biophys. Acta 1588 (1): 41–7. 2002. doi:10.1016/s0925-4439(02)00114-x. PMID 12379312.
- "Protein-protein interactions between large proteins: two-hybrid screening using a functionally classified library composed of long cDNAs.". Genome Res. 12 (11): 1773–84. 2003. doi:10.1101/gr.406902. PMID 12421765.
External links
- NFAT5+protein,+human at the US National Library of Medicine Medical Subject Headings (MeSH)
- Overview of all the structural information available in the PDB for UniProt: O94916 (Nuclear factor of activated T-cells 5) at the PDBe-KB.
This article incorporates text from the United States National Library of Medicine, which is in the public domain.
 |

