Biology:Basic helix-loop-helix
| basic helix-loop-helix DNA-binding domain | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
 | |||||||||||
| Identifiers | |||||||||||
| Symbol | bHLH | ||||||||||
| Pfam | PF00010 | ||||||||||
| InterPro | IPR001092 | ||||||||||
| SMART | SM00353 | ||||||||||
| PROSITE | PDOC00038 | ||||||||||
| SCOP2 | 1mdy / SCOPe / SUPFAM | ||||||||||
| CDD | cd00083 | ||||||||||
| |||||||||||
A basic helix-loop-helix (bHLH) is a protein structural motif that characterizes one of the largest families of dimerizing transcription factors.[2][3][4][5]
bHLH transcription factors are often important in development or cell activity. For one, BMAL1-Clock is a core transcription complex in the molecular circadian clock. Other genes, like c-Myc and HIF-1, have been linked to cancer due to their effects on cell growth and metabolism.
Structure
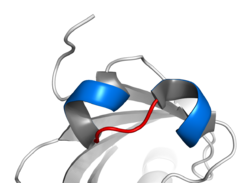
The motif is characterized by two α-helices connected by a loop. In general, transcription factors including this domain are dimeric, each with one helix containing basic amino acid residues that facilitate DNA binding.[6] In general, one helix is smaller, and, due to the flexibility of the loop, allows dimerization by folding and packing against another helix. The larger helix typically contains the DNA-binding regions. bHLH proteins typically bind to a consensus sequence called an E-box, CANNTG.[7] The canonical E-box is CACGTG (palindromic), however some bHLH transcription factors, notably those of the bHLH-PAS family, bind to related non-palindromic sequences, which are similar to the E-box. bHLH TFs may homodimerize or heterodimerize with other bHLH TFs and form a large variety of dimers, each one with specific functions.[8]
Examples
A phylogenetic analysis suggested that bHLH proteins fall into 6 major groups, indicated by letters A through F. [9] Examples of transcription factors containing a bHLH include:
Group A
- MyoD
- Myf5
- Beta2/NeuroD1
- Scl, also known as Tal1
- proneural bHLH genes like p-CaMKII, and pSer(336)NeuroD.
- Neurogenins
Group B
Group C
These proteins contain two additional PAS domains after the bHLH domain.
Group D
Group E
Group F
These proteins contain an additional COE domain
- EBF1
Regulation
Since many bHLH transcription factors are heterodimeric,[8] their activity is often highly regulated by the dimerization of the subunits. One subunit's expression or availability is often controlled, whereas the other subunit is constitutively expressed. Many of the known regulatory proteins, such as the Drosophila extramacrochaetae protein, have the helix-loop-helix structure but lack the basic region, making them unable to bind to DNA on their own. They are, however, able to form heterodimers with proteins that have the bHLH structure, and inactivate their abilities as transcription factors.[10]
History
- 1989: Murre et al. showed that dimers of various bHLH proteins bind to a short DNA motif (later called E-Box).[11] This E-box consists of the DNA sequence CANNTG, where N can be any nucleotide.[7]
- 1994: Harrison's[12] and Pabo's[13] groups crystallize bHLH proteins bound to E-boxes, demonstrating that the parallel 4-helix bundle motif loop orients the basic sequences to interact with specific nucleotides in the major groove of the E-box.
- 1994: Wharton et al. identified asymmetric E-boxes bound by a subset of bHLH proteins with PAS domains (bHLH-PAS proteins), including Single-minded (Sim) and the aromatic hydrocarbon receptor.[14]
- 1995: Semenza's group identifies hypoxia-inducible factor (HIF) as a bHLH-PAS heterodimer that binds a related asymmetric E-box.[15]
- 2009: Grove, De Masi et al., identified novel short DNA motifs, bound by a subset of bHLH proteins, which they defined as "E-box-like sequences". These are in the form of CAYRMK, where Y stands for C or T, R is A or G, M is A or C and K is G or T.[16]
Human proteins with helix-loop-helix DNA-binding domain
AHR; AHRR; ARNT; ARNT2; ARNTL; ARNTL2; ASCL1; ASCL2; ASCL3; ASCL4; ATOH1; ATOH7; ATOH8; BHLHB2; BHLHB3; BHLHB4; BHLHB5; BHLHB8; CLOCK; EPAS1; FERD3L; FIGLA; HAND1; HAND2; HES1; HES2; HES3; HES4; HES5; HES6; HES7; HEY1; HEY2; HIF1A; ID1; ID2; ID3; ID4; KIAA2018; LYL1; MASH1; MATH2; MAX; MESP1; MESP2; MIST1; MITF; MLX; MLXIP; MLXIPL; MNT; MSC; MSGN1; MXD1; MXD3; MXD4; MXI1; MYC; MYCL1; MYCL2; MYCN; MYF5; MYF6; MYOD1; MYOG; NCOA1; NCOA3; NEUROD1; NEUROD2; NEUROD4; NEUROD6; NEUROG1; NEUROG2; NEUROG3; NHLH1; NHLH2; NPAS1; NPAS2; NPAS3; NPAS4; OAF1; OLIG1; OLIG2; OLIG3; PTF1A; SCL; SCXB; SIM1; SIM2; SOHLH1; SOHLH2; SREBF1; SREBF2; TAL1; TAL2; TCF12; TCF15; TCF21; TCF3; TCF4; TCFL5; TFAP4; TFE3; TFEB; TFEC; TWIST1; TWIST2; USF1; USF2;
References
- ↑ PDB: 1x0o; "Structural basis of ARNT PAS-B dimerization: use of a common beta-sheet interface for hetero- and homodimerization". J. Mol. Biol. 353 (3): 664–77. October 2005. doi:10.1016/j.jmb.2005.08.043. PMID 16181639.
- ↑ "Structure and function of helix-loop-helix proteins". Biochim. Biophys. Acta 1218 (2): 129–35. June 1994. doi:10.1016/0167-4781(94)90001-9. PMID 8018712.
- ↑ "Transcription factors 2: helix-loop-helix". Protein Profile 2 (6): 621–702. 1995. PMID 7553065.
- ↑ "Helix-loop-helix proteins: regulators of transcription in eucaryotic organisms". Mol. Cell. Biol. 20 (2): 429–40. January 2000. doi:10.1128/MCB.20.2.429-440.2000. PMID 10611221.
- ↑ Amoutzias, Grigoris D.; Robertson, David L.; Van de Peer, Yves; Oliver, Stephen G. (2008-05-01). "Choose your partners: dimerization in eukaryotic transcription factors". Trends in Biochemical Sciences 33 (5): 220–229. doi:10.1016/j.tibs.2008.02.002. ISSN 0968-0004. PMID 18406148.
- ↑ Lawrence Zipursky; Arnold Berk; Monty Krieger; Darnell, James E.; Lodish, Harvey F.; Kaiser, Chris; Matthew P Scott; Matsudaira, Paul T. (2003-08-22). McGill Lodish 5E Package - Molecular Cell Biology & McGill Activation Code. San Francisco: W. H. Freeman. ISBN 0-7167-8635-4.
- ↑ 7.0 7.1 "Basic helix-loop-helix proteins can act at the E-box within the serum response element of the c-fos promoter to influence hormone-induced promoter activation in Sertoli cells". Mol. Endocrinol. 13 (5): 774–86. 1999. doi:10.1210/mend.13.5.0271. PMID 10319327.
- ↑ 8.0 8.1 Amoutzias, Gregory D.; Robertson, David L.; Oliver, Stephen G.; Bornberg-Bauer, Erich (2004-03-01). "Convergent evolution of gene networks by single-gene duplications in higher eukaryotes". EMBO Reports 5 (3): 274–279. doi:10.1038/sj.embor.7400096. ISSN 1469-221X. PMID 14968135.
- ↑ Ledent, V; Paquet, O; Vervoort, M (2002). "Phylogenetic analysis of the human basic helix-loop-helix proteins.". Genome Biology 3 (6): research0030.1. doi:10.1186/gb-2002-3-6-research0030. PMID 12093377.
- ↑ "Regulation of scute function by extramacrochaete in vitro and in vivo". Development 120 (12): 3595–603. 1994. PMID 7821225.
- ↑ Murre C; McCaw PS; Vaessin H et al. (1989). "Interactions between heterologous helix-loop-helix proteins generate complexes that bind specifically to a common DNA sequence". Cell 58 (3): 537–44. doi:10.1016/0092-8674(89)90434-0. PMID 2503252.
- ↑ "Crystal structure of transcription factor E47: E-box recognition by a basic region helix-loop-helix dimer". Genes Dev. 8 (8): 970–80. April 1994. doi:10.1101/gad.8.8.970. PMID 7926781.
- ↑ "Crystal structure of MyoD bHLH domain-DNA complex: perspectives on DNA recognition and implications for transcriptional activation". Cell 77 (3): 451–9. May 1994. doi:10.1016/0092-8674(94)90159-7. PMID 8181063.
- ↑ "Control of CNS midline transcription by asymmetric E-box-like elements: similarity to xenobiotic responsive regulation". Development 120 (12): 3563–9. December 1994. PMID 7821222.
- ↑ "Hypoxia-inducible factor 1 is a basic helix-loop-helix-PAS heterodimer regulated by cellular O2 tension". Proc. Natl. Acad. Sci. U.S.A. 92 (12): 5510–4. June 1995. doi:10.1073/pnas.92.12.5510. PMID 7539918.
- ↑ Grove C; De Masi F et al. (2009). "A multiparameter network reveals extensive divergence between C. elegans bHLH transcription factors". Cell 138 (2): 314–27. doi:10.1016/j.cell.2009.04.058. PMID 19632181.
External links
- PDOC00038 in PROSITE
- Basic+Helix-Loop-Helix+Transcription+Factors at the US National Library of Medicine Medical Subject Headings (MeSH)
- bHLH family at PlantTFDB:Plant Transcription Factor Database
